Abstract
We characterized the complete chloroplast genome of Alsophila gigantea (Cyatheaceae), an ornamental and CITES giant tree fern. Its genome size is 161,679 bp in length, containing a large single copy (LSC) of 92,315 bp and a small single copy (SSC) of 21,702 bp separated by a pair of inverted repeat regions (IRs) of 23,831 bp. The overall GC contents of the chloroplast genome are 41.4%. A total of 133 genes are annotated, comprising 89 protein-coding genes, 33 tRNA, eight rRNA, and three pseudogenes. ML phylogenetic tree indicates that A. gigantea is sister to A. podophylla.
Alsophila gigantea, known as Cyathea gigantea, is a giant tree fern belonging to Cyatheaceae (Zhang and Nishida Citation2013). Its erect trunk is 2–5 meter tall with remnants of older leaf bases. The stipe is covered with dark brown and glossy scales. The species mainly grows in moist open areas in dense forests at an altitude up to 1200 m in India, Sri Lanka, Burma, Thailand, Laos, Vietnam, and southern China (Zhang and Nishida Citation2013). Similar to other species of Alsophila, A. gigantea is not only used as a source of starch, but also its frond is used for growing epiphytic orchids (Kiran et al. Citation2012). As an ornamental fern with a loose crown of leaves, it has been included in Appendix II of CITES (Citation2017) due to illegal collection. In addition, A. gigantea forms a complex with other morphologically similar species of Alsophila such as A. glabra, A. podophylla, and A. subdubia (Large and Braggins Citation2004). Hence, acquirement of its whole chloroplast genome sequence will contribute to determine the relationship between these taxa.
In this study, the sample was collected from South China Botanical Garden, Chinese Academy of Sciences (23°11′3.56″N, 113°21′43.28″E) and deposited in Herbarium of Sun Yat-sen University (SYS; voucher: SS Liu 201618). Total genomic DNA was extracted from young leaves using Tiangen Plant Genomic DNA Kit (Tiangen Biotech Co., Beijing, China) and further broken into approximately 300 bp fragments. After ligated barcode adaptors, a paired-end library was constructed and sequenced using Illumina Hiseq 2500 platform (Illumina Inc., San Diego, CA).
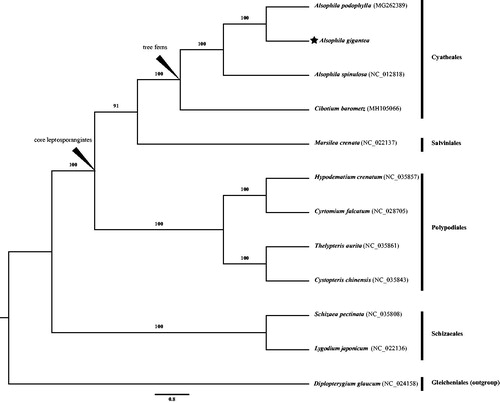
In total, 12,053,058 raw reads were obtained. After low-quality reads and adapters were trimmed by Trimmomatic (Bolger et al. Citation2014), remaining 11,109,827 high-quality clean reads were de novo assembled to complete chloroplast genome by Velvet v1.2.07 (Zerbino and Birney Citation2008). Gaps were filled by PCR-based methods. The genes were annotated using DOGMA (Wyman et al. Citation2004) and tRNAscan-SE (Schattner et al. Citation2005) with the default configuration and were further manually curated against the plastome of A. spinulosa (NC_012818). The complete chloroplast genome sequences of A. gigantea were aligned with 11 species from leptosporangiates including Diplopterygium glaucum as an outgroup using MAFFT v7.311 (Katoh and Standley Citation2013). A maximum likelihood (ML) phylogenetic tree was constructed using raxmlGUI version 1.5b (https://sourceforge.net/projects/raxmlgui/) (Silvestro and Michalak Citation2012) with 1000 bootstrap replicates.
The plastome of A. gigantea was found to possess a total length 161,679 bp with 41.4% GC content (Genbank accession number: MH603068). It is a typical quadripartite structure containing a large single copy (LSC) of 92,315 bp and a small single copy (SSC) of 21,702 bp separated by a pair of inverted repeat regions (IRs) of 23,831 bp. A total of 133 genes were annotated, comprising 89 protein-coding genes, 33 tRNA, eight rRNA, and three pseudogenes. Among them, 13 have two copies. Except three genes with two introns (ycf3, clpP, and rps12), 14 genes harbor an intron (ndhB, rps16, atpF, rpoC1, petB, petD, rpl16, rpl2, ndhA, trnG-UCC, trnV-UAC, trnA-UGC, trnI-GAU, and trnL-CAA). ML phylogenetic tree indicates that A. gigantea is sister to A. podophylla with high bootstrap support value ().
Disclosure statement
The authors declare no conflict of interests. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Large MF, Braggins JE. 2004. Tree ferns. Portland (OR): Timber Press; p. 136.
- CITES. 2017. Convention on International Trade in Endangered Species of Wild Fauna and Flora. Appendices I, II, and III. [accessed 2018 Jun 7]. https://cites.org/eng/app/appendices.php
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kiran PM, Raju AV, Rao BG. 2012. Investigation of hepatoprotective activity of Cyathea gigantea (Wall. ex. Hook.) leaves against paracetamol-induced hepatotoxicity in rats. Asian Pac J Trop Biomed. 2:352–356.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Silvestro D, Michalak I. 2012. RaxmlGUI: a graphical front-end for RaxML. Org Divers Evol. 12:335–337.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhang XC, Nishida H. 2013. Cyatheaceae. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol. 2–3 (Pteridophytes). Beijing: Science Press; p. 134–138.