Abstract
The complete chloroplast genome of Rosa banksiae var. normalis was reconstructed by reference-based assembly using Illumina paired-end data. The assembled plastome is 156,575 base pairs (bp) in length, including a pair of inverted repeat regions (IRs) of 26,008 bp each, a large single copy region (LSC) of 85,792 bp, and a small single copy region (SSC) of 18,767 bp. A total of 129 genes were predicted from the chloroplast genome, including 84 protein coding genes, 37 tRNA genes and 8 rRNA genes. The overall GC content of Rosa banksiae var. normalis chloroplast genome was 37.22%. Phylogenetic analysis with several reported chloroplast genomes showed that Rosa banksiae var. normalis is closely clustered with Rosa odorata var. gigantea, Rosa multiflora, Rosa chinensis var. spontanea, and Rosa roxburghii. The complete chloroplast genome of Rosa banksiae var. normalis provides new insight into Rosaceae evolutionary and genomic studies.
Rosa banksiae var. normalis, with common name Lady Banks’ rose, is a species of flowering plant in the rose family, native to central and western China, at altitudes of 500–2,200 m. It has been widely cultivated for its high ornamental value. In addition, the root of Rosa banksiae var. normalis is a well-known traditional Chinese medicine. However, the phylogenetic position of Rosa banksiae var. normalis in the genus Rosa is still unclear. In this study, we first reported the complete chloroplast (cp) genome of Rosa banksiae var. normalis.
The mature and healthy leaves of a single individual of Rosa banksiae var. normalis was sampled from the field of Midu county in western Yunnan, SW China. The voucher specimen was deposited in the Herbarium of Chengdu Institute of Biology (CDBI). The total genomic DNA was extracted from silica gel dried leaves using a modified CTAB method (Doyle and Doyle Citation1987) and sequenced based on the Illumina pair-end technology. The filtered reads were assembled using the program NOVOPlasty (Dierckxsens et al. Citation2017) with complete chloroplast genome of Rosa multiflora as the reference (GenBank accession no. MG893867). The assembled chloroplast genome was annotated using Plann (Huang and Cronk Citation2015) and the annotation was corrected using Geneious (Kearse et al. Citation2012). To examine the phylogenetic position of Rosa banksiae var. normalis in Rosaceae, a maximum likelihood (ML) tree was constructed using RAxML v8.2.11 (Stamatakis Citation2006) with 1000 bootstrap replicates based on the alignments created by the MAFFT (Katoh and Standley Citation2013) using already published plastid genomes.
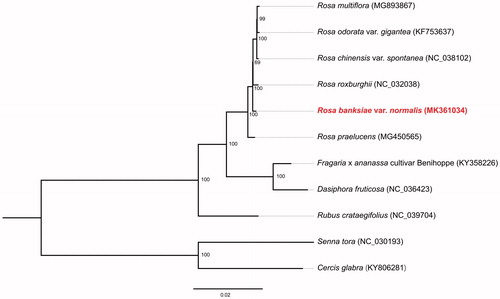
The complete cp genome of Rosa banksiae var. normalis (Genbank accession no. MK361034) was a circular molecular genome with a size of 156,575 bp in length, which presented a typical quadripartite structure containing two inverted repeat (IR) regions of 26,008 bp separated by the large single copy (LSC) region of 85,792 bp and small single copy (SSC) region of 18,767 bp. The cp genome consists of 129 genes including 84 protein coding genes, 37 tRNA genes, and 8 rRNA genes. The overall GC content was about 37.22%. Our phylogenetic results showed that Rosa banksiae var. normalis is located in the genus Rosa and is closely clustered with Rosa odorata var. gigantea, Rosa multiflora, Rosa chinensis var. spontanea, and Rosa roxburghii (100% bootstrap support) (). This complete cp genome can be further used for population genomic studies, phylogenetic analyses, genetic engineering studies of Rosaceae.
Acknowledgements
The authors thank Zhengzhi Jiang from Suzhou Huaguanyuanchuang Horticultural and Technology Ltd. for the help of plant material collection.
Disclosure statement
None of the co-authors have any conflict of interest to declare. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Huang DI, Cronk QCB. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.