Abstract
Camellia japonica L. is an ornamental species due to its beautiful red flowers blooming in winter. Here, we completed the chloroplast genome of C. japonica isolated in Soyeonpyeongdo located at the northernmost in South Korea. Its length is 156,971 bp long and has four subregions: 86,673 bp of a large single copy (LSC) and 18,394 bp of small single copy (SSC) regions are separated by 25,952 bp of inverted repeat (IR) regions including 135 genes (91 protein-coding genes, 8 rRNAs, and 36 tRNAs). The overall GC content of this chloroplast genome is 37.3% and those in the LSC, SSC, and IR regions are 35.3%, 30.5%, and 43.0%, respectively. Twenty-five single nucleotide polymorphisms (SNPs) and two insertions and deletions (INDELs) are identified against Korean C. japonica isolated in Jeju island, while 78 SNPs and 643 INDELs are found against Chinese C. japonica. Phylogenetic trees show that Camellia chekiangoleosa is clustered with Korean C. japonica.
Genus Camellia L. containing around 200 species (Ye Citation1997), is evergreen shrubs and distributes in eastern and southern Asia from the Himalayas east to Japan and Indonesia (Kondo Citation1977; Ming Citation2000). Owing to its beautiful flowers, more than 3,000 hybrids have been developed. Camellia japonica L. is one of the best-known species in genus Camellia called as the rose of winter. However, C. japonica cannot survive below −10 °C (Byers Citation1999). Northern most habitats of C. japonica in Korea is Soyeonpyeongdo of which average lowest temperature in January was −10.7 °C between 2005 and 2009 (Kim et al. Citation1992) and −14 °C in 2017. It is similar environment to the northern most habitats in Japan (Wheeler et al. Citation2015), indicating that C. japonica in Soyeonpyeongdo may have resistance against coldness.
To understand its genetic background, total DNA of C. japonica collected in Soyeonpyeongdo, Incheon, Korea (N37°36′ 46.45″ and E125°42′44.66″; Voucher in InfoBoss Cyber Herbarium (IN); Y. Kim, IB-00581) was extracted from fresh leaves by using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Genome was sequenced using HiSeqX at Macrogen Inc., Korea, and de novo assembly was done by Velvet 1.2.10 (Zerbino and Birney Citation2008) and SOAPGapCloser 1.12 (Zhao et al. Citation2011) and confirmed by BWA 0.7.17 (Li Citation2013) and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.0.5 (Biomatters Ltd., Auckland, New Zealand) was used for chloroplast genome annotation based on C. japonica chloroplast complete genome (KU951523; Kim et al. Citation2017).
The chloroplast genome of C. japonica (MK353210) is 156,971 bp and has four subregions: 86,673 bp of a large single copy (LSC) and 18,394 bp of small single copy (SSC) regions are separated by 25,952 bp of the inverted repeat (IR). It contains 135 genes (91 protein-coding genes, 8 rRNAs, and 36 tRNAs); 19 genes (8 protein-coding genes, 4 rRNAs, and 7 tRNAs) are duplicated in IR regions. The overall GC content of C. japonica is 37.3% and those in the LSC, SSC, and IR regions are 35.3%, 30.5%, and 43.0%, respectively.
Based on alignment with Korean C. japonica, chloroplast genome isolated in Jeju Island, Korea (Kim et al. Citation2017), 25 single nucleotide polymorphisms (SNPs) and two insertions and deletions (INDELs) are identified. One deletion increases two amino acids of the psbL gene, two non-synonymous SNPs, and one synonymous SNP are on psbE, and four SNPs are in trnP. In addition, 78 SNPs and 643 INDELs are found against Chinese C. japonica (NC_036830).
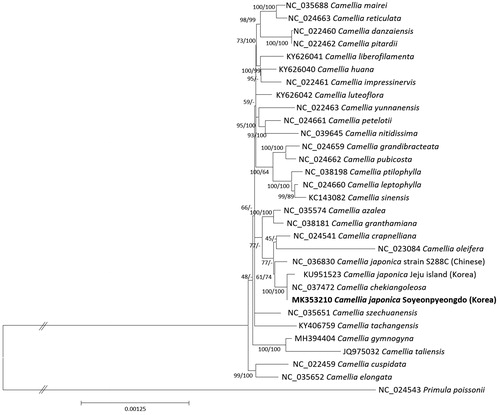
Thirty chloroplast genomes of Camellia including three C. japonica chloroplasts (Kim et al. Citation2017) and one outgroup Theaceae chloroplast genome were used for constructing neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenic trees using MEGA X (Kumar et al. Citation2018) after aligning whole chloroplast genome sequences by MAFFT 7.388 (Katoh and Standley Citation2013). Phylogenetic trees show that two Korean C. japonica are clustered with Cameliia chekiangoleosa which is distributed in the subtropical region of China but fairly hard to endure −10 °C (Grimshaw and Bayton Citation2009), hence Chinese C. japonica is placed outside of this clade ().
Figure 1 Neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenetic tree of 30 Camellia and one Primula chloroplast genomes: Camellia japonica (MK353210 in this study, KU951523, and NC_036830), Camellila chekiangoleosa (NC_037472), Camellia crapnelliana (NC_024541), Camellia oleifera (NC_024541), Camellia granthamiana (NC_038181), Camellia azalea (NC_035574), Camellia sinensis (NC_024541), Camellia letophylla (NC_024541), Camellia ptilophylla (NC_038198), Camellia pubicosta (NC_024541), Camellia grandibracteata (NC_024541), Camellia nitidissima (NC_039645), Camellia petelotii (NC_024541), Camellia yunnanensis (NC_024541), Camellia luteoflora (KY626042), Camellia impressinevis (NC_024541), Camellia huana (KY626040), Camellia liberofilamenta (KY626041), Camellia pitardii (NC_024541), Camellia danzaiensis (NC_024541), Camellia reticulata (NC_024541), Camellia mairei (NC_035688), Camellia szechuanensis (NC_035651), Camellia tachangensis (KY406759), Camellia gymnogyna (MH394404), Camellia tallensis (JQ975032), Camellia cuspidata (NC_024541), Camellia elongate (NC_035652), and Primula polssonii (NC_024543). The numbers above branches indicate bootstrap support values of neighbor joining and maximum likelihood phylogenetic tree, respectively.
Disclosure statement
The authors declare that they have no competing interests.
Additional information
Funding
References
- Byers RD. 1999. Reaching out: a university botanical garden builds long-distance relationships. HortTechnology. 9:573–576.
- Grimshaw J, Bayton R. 2009. New trees: recent introductions to cultivation. Royal Botanic Gardens.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kim S-H, Cho CH, Yang M, Kim S-C. 2017. The complete chloroplast genome sequence of the Japanese Camellia (Camellia japonica L.). Mitochondr DNA B. 2:583–584.
- Kim T-W, Chun S-H, Kang G-H. 1992. Floristic study of Baekryong and Yeonpyong Island within Ongjin Gun, Kyonggi Province. Seoul National Univ Col of Agric Rec. 12:30–46.
- Kondo K. 1977. Chromosome numbers in the genus Camellia. Biotropica. 9:86–94.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Ming T. 2000. Monograph of the genus Camellia. Kunming, China: Yunnan Science and Technology Press; p. 128–134.
- Wheeler L, Su M, Rivers MC. 2015. Camellia japonica. The IUCN Red List of Threatened Species 2015: e.T62054114A62054131. [accessed 2018 December 26].
- Ye C. 1997. Classification in the genus Camellia L. American Camellia Society: American Camellia Yearbook.9-23.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12:S2.
