Abstract
Vischeria stellata SAG 33.83 is an edaphic oleaginous microalga which is able to accumulate massive storage triacylglycerols (TAG) with nutritionally valuable palmitoleic acid (PA) and membrane lipids with substantial amounts of eicosapentaenoic acid (EPA). The features of the fatty acids metabolism make such alga highly attractive for biotechnological applications. This study first determined and assembled the complete chloroplast genome of V. stellata SAG 33.83 by Illumina sequencing data. It was found that the circular genome made up of 126,824 bp with 32.6% GC content, including 142 protein-coding genes (PCGs), 29 transfer RNA genes (tRNAs), 6 ribosomal RNA genes (rRNAs), and 1 transfer-messenger RNA (tmRNA). The chloroplastic genome composition and structure of V. stellata SAG 33.83 were almost identical to other species of Eustigmatophyceae. Nucleotide of stramenopile species was aligned with 28,824 bp including 54 genes in series as phylogenetic analysis, which demonstrated a close relationship between V. stellata SAG 33.83 and Vischeria sp. CAUP Q 202.
Vischeria stellata SAG 33.83, belonging to Eustigmatophyceae, was originally isolated from the soil of mountain Isle Lavsa, Dalmatia, Europe (43.7521°N, 15.3695°W) and deposited in the Culture Collection of Algae at Göttingen University (SAG). It was obtained from SAG as our experimental material. This species can accumulate large amounts of lipids and β-carotene, which can reach up to 66.8 and 2.0% of dry weight, respectively (Gao et al. Citation2016; Wang et al. Citation2018). V. stellata SAG 33.83 was an oleaginous microalga which had ecologically and economically important value and considered to be a potential producer of biofuels and nutraceuticals.
Previous researches about V. stellata were focused on morphological characteristics and optimization of culture conditions (Santos and Leedale Citation1991; Gärtner et al. Citation2012; Gao et al. Citation2016). Efforts to explore and clarify V. stellata intrinsic values were hindered by a lack of knowledge of its genetic system, phylogenetic information, and metabolic network structure. To reveal these fundamental issues, the complete mitochondrial genome of V. stellata SAG 33.83 had been sequenced (Huang et al. Citation2019). However, no information of chloroplastic genome had been reported. Through this study, the complete chloroplast genome of the V. stellata SAG 33.83 was recovered through Illumina sequencing data and its genomic rofile and phyletic evolution were then analyzed.
The DNA and genomic libraries of V. stellata SAG 33.83 were acquired in our laboratory (Huang et al. Citation2019) and sequenced through Illumina HiSeq4000 platform by BGI Biotechnology Co. Ltd (Shenzhen, China). A total of 41.43 Mbp raw reads were obtained and trimmed with NGS QC Toolkit 2.3.3 (Patel and Jain Citation2012). The chloroplast genome assembly was performed by SPAdes 3.9.0 with a default value (Bankevich et al. Citation2012) and identified by comparing the sequence similarity to other eustigmatophycean chloroplast genomes. Finally, the annotation of the chloroplast genome was performed by using online server tools such as DOGMA, CpGAVAS, tRNAscan-SE, and RNAmmer 1.2 combined with manual correction.
The complete and annotated chloroplast genome sequence of V. stellate SAG 33.83 had been submitted to the GenBank database under the access number of MK212028. The complete chloroplast genome was a circular structure of 126,824 bp in length with a highly biased base composition (33.3% A, 16.8% C, 15.8% G, and 34.2% T) and GC content of 32.6%. A total of 178 genes were identified in this process, including 142 protein-coding genes (PCGs), 29 transfer RNA genes (tRNAs), 6 ribosomal RNA genes (rRNAs), and 1 transfer-messenger RNA (tmRNA).
In order to identify the phylogenetic relationship of V. stellata SAG 33.83 within the stramenopile, a maximum likelihood (ML) phylogenetic tree was conducted with MEGA7 (Kumar et al. Citation2016) using 28,824 bp collinear alignments (including 54 genes), which was calculated by HomBlocks (Bi et al. Citation2018) of 30 relative species of chloroplastic genome downloaded from NCBI and sequence of V. stellata SAG 33.83. As shown in the phylogenetic tree (), V. stellata SAG 33.83 was clustered in the Eustigmataceae clade and closely clustered with Vischeria sp. CAUP Q 202 (KX839261) (Yurchenko et al. Citation2016). Meanwhile, the phylogenetic tree showed that 12 species of Eustigmataceae clustered into one monophyletic clade, which was categorized into two different branches together with other six classes of stramenopiles, including Bacillariophyceae, Pelagophyceae, Raphidophyceae, Xanthophyceae, Phaeophyceae, and Chrysophyceae ().
Figure 1. Phylogenetic analysis of Vischeria stellata SAG 33.83. The maximum likelihood phylogenetic tree was inferred from the 28,824 bp collinear sequences of 54 genes in all 30 algae species, by the use of General Time Reversible and Gamma distributed with invariant sites substitution model(GTR + G+I). The significant level of the phylogenetic tree was determined by bootstrap testing with 1000 replications. Bootstrap support values were then shown in branches when they reached over 88%. GenBank accession numbers are shown in parentheses and the scale bar indicates 0.10 substitutions per nucleotide position.
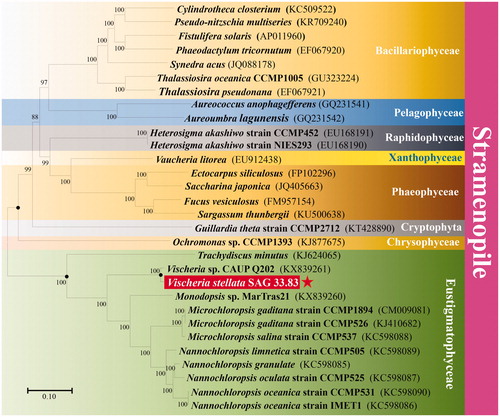
According to endosymbiosis theory, the chloroplast of stramenopile was originated from engulfment of photosynthetic eukaryotic algae, which is secondary endosymbiosis (Curtis et al. Citation2012; Suo et al. Citation2018). Therefore, it was assumed that there were different types of engulfed photosynthetic organisms and different durations occurring between eustigmatophycean species and other stramenopile species. These results further confirmed that Eustigmatophyceae had a unique and complex evolutionary history compared to other stramenopile species ().
It was concluded that the complete chloroplast genome sequence obtained in this study would be useful for investigating the phylogenetic history of V. stellata, Eustigmatophycean species, and endosymbiosis.
Disclosure statement
No potential conflict of interest was reported by the authors. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SL, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Bi G, Mao Y, Xing Q, Cao M. 2018. Homblocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110:18–22.
- Curtis BA, Tanifuji G, Burki F, Gruber A, Irimia M, Maruyama S, Arias MC, Ball SG, Gile GH, Hirakawa Y, et al. 2012. Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature. 492:59–65.
- Gao B, Yang J, Lei X, Xia S, Li A, Zhang C. 2016. Characterization of cell structural change, growth, lipid accumulation, and pigment profile of a novel oleaginous microalga, Vischeria stellata (Eustigmatophyceae), cultured with different initial nitrate supplies. J Appl Phycol. 28:821–830.
- Gärtner G, Stoyneva MP, Uzunov BA, Mancheva AD, Ingolić E. 2012. Ultrastructure of vegetative cells and autospores of an aerophytic strain of Vischeria stellata (Chodat ex Poulton) Pascher (Eustigmatophyceae) from Bulgaria. Fottea. 12:273–280.
- Huang L, Gao B, Wang F, Zhang C. 2019. The complete mitochondrial genome of an oleaginous microalga Vischeria stellata strain SAG 33.83 (Eustigmatophyceae). Mitochondr DNA B. 4:301–302.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7:e30619.
- Santos LM, Leedale GF. 1991. Vischeria stellata (Eustigmatophyceae): ultrastructure of the zoospores, with special reference to the flagellar apparatus. Protoplasma. 164:160–167.
- Yurchenko T, Ševčíková T, Strnad H, Butenko A, Eliáš M. 2016. The plastid genome of some eustigmatophyte algae harbours a bacteria-derived six-gene cluster for biosynthesis of a novel secondary metabolite. Open Biol. 6:1–22.
- Suo F, Ma Y, Manzilamu Z, Wang Y, Huang L. 2018. The complete mitochondrial genome of a transitional form in secondary endosymbiotic Cryptophyte algae Guillardia theta strain CCMP2712. Mitochondr DNA B. 3:1304–1305.
- Wang F, Gao B, Huang L, Su M, Dai C, Zhang C. 2018. Evaluation of oleaginous eustigmatophycean microalgae as potential biorefinery feedstock for the production of palmitoleic acid and biodiesel. Bioresour Technol. 270:30–37.
