Abstract
We describe the complete mitochondrial genome of the nematode-trapping fungus Arthrobotrys musiformis. In this study, the complete mitochondrial genome of A. musiformis was sequenced. This mitogenome is a circular molecule of 179,060 bp in length. Gene annotation showed that 14 protein-coding genes, 16 transfer RNAs, and 2 ribosomal RNAs, all located on the same strand. The evolutionary relationships between A. musiformis and other representative ascomycetes were revealed based on the sequences at the 14 concatenated mitochondrial protein-coding genes.
Arthrobotrys musiformis is an important member of many nematode-trapping fungi and its hyphae can be specialized to form a three-dimensional network, which can trap vermiform nematodes or hyphal tips to attack nematode eggs and cysts before penetration of the nematode cuticle, invasion, and digestion (Nordbring-Hertz et al. Citation2011). As early as the 1930s, Linford had used the species of A. musiformis to control pineapple root-knot nematodes and achieved certain effects, it can be seen that this species has already demonstrated the potential biological control of animal and plant parasites (Bi et al. Citation2000). However, little is known about its mitogenome. Here, we report the complete mitogenome of A. musiformis and investigate the phylogenetic relationship with other related species based on mitochondrial proteins.
The mitogenome was extracted from the whole genome sequence of a pure culture of strain YMF1.03721 (preserved in the State Key Laboratory for Conservation and Utilization of Bio-Resources in Yunnan). We sequenced it using second-generation sequencing technology and assembled it using A5-miseq v20150522 (Coil et al. Citation2015) and SPAdesv3.9.0 (Bankevich et al. Citation2012). The mitogenome is automatically annotated by MFannot tool (http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl) and GLIMMER (https://www.ncbi.nlm.nih.gov/genomes/MICROBES/glimmer_3.cgi) based on the old mitochondrial genetic code 4. tRNAs of A. musiformis were annotated using tRNAscan-SE (Schattner et al. Citation2005; Lowe and Chan Citation2016). All ORFs were searched and identified by ORF Finder.
The complete mitogenome of A. musiformis is a closed circular molecule of 179,060 bp in length with A content of 38.66%, G content of 13.47%, C content of 11.11%, T content of 36.76%, including 14 mitochondrial protein-coding (PCGs) genes, 16 tRNA genes which codes 13 standard amino acids, and 2 rRNA genes. Protein-encoding genes include one apocytochrome b (cob), three cytochrome oxidase subunits (cox1, cox2, and cox3), three ATP synthase subunits (atp6, atp8, and atp9), and seven NADH dehydrogenase subunits (nad1, nad2, nad3, nad4, nad4L, nad5, and nad6). It is worth noting that 12 of the 14 encoded proteins use ATG as a start codon, ATP8 started with TTG, and NADH1 began with ATA, all of 14 PCGs used TAA as a stop codon. The all predicted genes totally possessed 42 introns, but none of the tRNAs possessed introns. All tRNA genes are encoded on the sense strand and it does not contain the seven amino acids Cysteine (Cys), Proline (Pro), Isoleucine (Iie), Leucine (Leu), Tryptophan (Trp), Glycine (Gly), Aspartic acid (Asp).
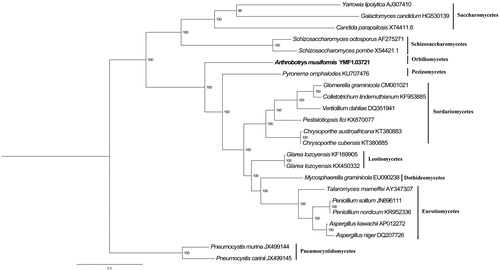
Phylogenetic analysis of 14 mitochondrial protein-coding genes of A. musiformis and other 22 species in Ascomycota was performed by Bayesian inference (BI). The best substitution model was analyzed by ProtTest 3.0 (Best model according to AIC: LG + G+F). As shown in . A. musiformis belongs to Orbiliomycetes closely related to Pyronema omphalodes. Pneumocystis which belongs to Pneumocystidomycetes was taken as an outgroup. The phylogenetic relationship using mitochondrial proteins were in accordance with those constructed based on sequences of nuclear genes (Sugiyama et al. Citation2006). The Genbank Accession number for Arthrobotrys musiformis is MK547645.
Figure 1. Phylogenetic relationships among 23 ascomycetes based on 14 conserved mitochondrial protein-coding genes. Phylogenetic analysis of 23 species of Agaricomycotina constructed using the Bayesian inference method as implemented in MrBayes based on concatenated amino acid sequences of 14 mitochondrial protein-coding genes. The following 14 mitochondrial protein-coding genes were concatenated: atp6, atp8, atp9, cytb, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad4L, nad5 and nad6. The concatenated amino acid sequences were aligned using Clustal X. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000,000 replicates) were shown next to the branches.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich A A, Dvorkin M, Kulikov A S, Lesin V M, Nikolenko S I, Pham S, Prjibelski A D, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bi TJ, Zhang KQ, Li WP. 2000. The resource survey of predacious fungi in Yunnan Province. J Yunnan Univ. 22:71–75.
- Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 31(4):587–589.
- Lowe TM, Chan PP. 2016. tRNAscan-SE on-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44:W54–W57.
- Nordbring-Hertz B, Jansson HB, Tunlid A. 2011. Nematophagous fungi. In: Pettis G, editor. eLS. Chichester: Wiley.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Sugiyama J, Hosaka K, Suh S-O. 2006. Early diverging Ascomycota: phylogenetic divergence and related evolutionary enigmas. Mycologia. 98:996–1005.
