Abstract
The complete cp genome of Salvia przewalskii was 152,678 bp in length, including a large single copy region (LSC) of 84,102 bp, a small single copy region (SSC) of 17,638 bp and a pair of inverted repeats (IRs) of 25,469 bp. The genome contained 131 genes, including 86 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The overall GC content of this genome was 38.0%, with the corresponding values of LSC, SSC, and IR regions being 36.2, 31.9, and 43.2%, respectively. Further, the phylogenomic analysis of Mentheae strongly supported the sister relationship of Salvia przewalskii and Salvia miltiorrhiza.
Salvia przewalskii Maxim. (Gansu sage) is a perennial herb of the genus Salvia (Lamiaceae), which is endemic to the Chinese provinces of Gansu, Hubei, Sichuan, Tibet, and Yunnan (Li and Ian Citation1994). It is widely known throughout its native habitat for its medicinal properties. The seeds of Salvia przewalskii can be used as a Chinese herbal medicine to promote blood circulation by removing blood stasis (Li et al. Citation2013). Salvia przewalskii attracts more and more researchers’ attention because of similar chemical composition with Salvia miltiorrhiza Bunge (Xu Citation2005). The roots of Salvia przewalskii has been used as a substitute for Salvia miltiorrhiza for many years (Weng et al. Citation2015). In recent years, Salvia miltiorrhiza will face the trend of short supply with the increasing of development and utilization of it (Fang et al. Citation2018). In order to better develop and utilize Gansu sage, we assembled and described the complete chloroplast genome sequence of Salvia przewalskii using the genome skimming sequencing method in this study.
Fresh leaves of Salvia przewalskii were collected from Deqin, Yunan. The voucher specimen (FW11193) was deposited in Herbarium, Kunming Institute of Botany, CAS (KUN). Total DNA was extracted under the modified CTAB method (Doyle and Doyle Citation1987). The genome skimming sequencing was performed on the Illumina HiSeq 2500 platform in Novogene Bioinformatics Technology Co., Ltd. (Beijing, China). We used CLC Genomics Workbench v.8.5.1 (CLC Bio, Aarhus, Denmark) to assemble the cp genome with Salvia miltiorrhiza for reference (Accession number NC_020431) (Qian et al. Citation2013). The annotation of the cp genome was developed through Dual Organellar Genome Annotator (DOGMA) (Wyman et al. Citation2004) and manually adjusted the position of start and stop codons. The tRNA genes were further confirmed by online tRNAscan-SE Search Service (Lowe and Chan Citation2016; http://lowelab.ucsc.edu/tRNAscan-SE/). The complete cp genome sequence was submitted to the GenBank under the accession number of MK344723. The circular map of the cp genome was drawn with the OGDRAW (Lohse et al. Citation2013; http://ogdraw.mpimp-golm.mpg.de/). The Salvia przewalskii cp genome was 152,678 bp in size, containing a large single copy region (LSC) of 84,102 bp and a small single copy region (SSC) of 17,638 bp separated by a pair of inverted repeats (IRs) of 25,469 bp. The genome had 131 genes, including 86 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The overall GC content of this genome was 38.0%, with the corresponding values of LSC, SSC, and IR regions being 36.2, 31.9, and 43.2%, respectively.
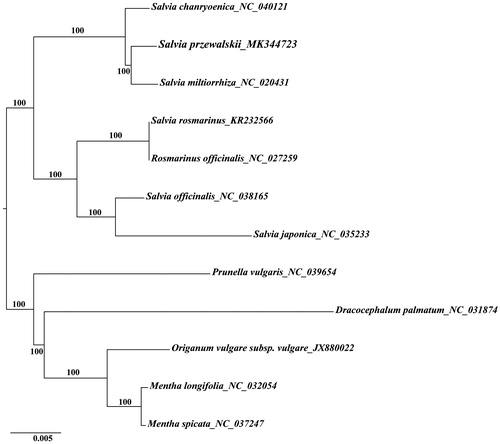
To detect the possibility of genomic data in the phylogenetic analysis, we downloaded all the complete cp genome sequences of Mentheae (Lamiaceae) available from GenBank and constructed the ML (Maximum Likelihood) phylogenetic tree with the GTR + G model. The phylogenetic results indicated that Salvia is not monophyletic, which is in accordance with previous studies (Sytsma and Drew Citation2011; Drew and Sytsma Citation2012). Further, Salvia przewalskii and Salvia miltiorrhiza formed a strongly-supported clade (), which was consistent with that they have similar pharmacological activities (Xue et al. Citation2000).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Drew BT, Sytsma KJ. 2012. Phylogenetics, biogeography, and staminal evolution in the tribe Mentheae (Lamiaceae). Am J Bot. 99:933–953.
- Fang WT, Deng AP, Ren ZL, Nan TG, Kang LP, Guo LP, Huang LQ, Zhan ZL. 2018. Research progress on quality evaluation of Salviae Miltiorrhizae Radix et Rhizoma (Danshen). J Chin Mater Med. 43:1077–1085.
- Li XW, Ian CH. 1994. Lamiaceae. In: Wu ZY, Raven PH, editors. Flora of China. Vol. 17. Beijing: Science Press; St. Louis: Missouri Botanical Garden Press; p. 50–299.
- Li MH, Li QQ, Liu YZ, Cui ZH, Zhang N, Huang LQ, Xiao PG. 2013. Pharmacophylogenetic study on plants of genus Salvia L. from China. Chin Herb Med. 5:164–181.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44:W54–W57.
- Qian J, Song JY, Gao HH, Zhu YJ, Xu J, Pang XH, Yao H, Sun C, Li XE, Li CY, et al. (2013) The complete chloroplast genome sequence of the medicinal plant Salvia miltiorrhiza. PLoS ONE: e57607.
- Sytsma KJ, Drew BT. 2011. Testing the monophyly and placement of Lepechinia in the tribe Mentheae (Lamiaceae). Syst Bot. 36:1038–1049.
- Weng YX, Liu ZH, Xiang HC. 2015. Pharmacognostic studies of Salvia przewalskii Maxim. J Pharm Res. 34:144–146.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Xu G. 2005. Study on the chemical constituents of five Salvia species [Dissertation]. Kunming: Kunming institute of Botany.
- Xue M, Shi YB, Cui Y, Zhang B, Luo YJ, Zhou ZT, Xia WJ, Zhao RC, Wang HQ. 2000. Study on the chemical constituents from Salvia przewalskii Maxim. Nat Prod Res Dev. 12:27–32.