Abstract
Here in this work, we have sequenced the complete chloroplast genome of Comastoma falcatum. The complete chloroplast genome size of C. falcatum is 151,423 bp with 81721 bp of LSC, 25727 bp of IRa/IRb and 18248 bp of SSC. The GC contents of its chloroplast genomes are 38.26%. Their chloroplast has a total of 133 unique genes, including 88 of protein-coding genes, 8 of rRNA genes, and 37 of tRNA genes. In the maximum parsimony phylogenetic tree, C. falcatum clustered with Swertia mussotii. This sequenced chloroplast genome of C. falcatum will help to further study the phylogenetic position of Comastoma.
Comastoma (Gentianaceae) is a small genus of 15 species distributed in Asia, Europe, and North America (Ho and James Citation1995). However, the evolutionary relationships among Comastoma species and its allied groups are still confused (Yuan and Küpfer Citation1995; Hagen and Kadereit Citation2001). There are different studies (Chassot et al. Citation2001; Hagen and Kadereit Citation2002; Brakefield Citation2006) about the genus but still with the question that whether the genus is monophyletic or not? In this study, we sequenced complete chloroplast genome of C. falcatum which will help to generate deep phylogeny of the genus.
Total genomic DNA of C. falcatum was extracted from leaf materials (about 100 mg) with a modified CTAB method (Doyle and Doyle Citation1987). The voucher specimen has collected at Maduo County in Qinghai Province (Accession No. Chensl-0120; Geographic coordinates 34°49′07″N, 99°02′33″E; Altitude 4680m) and deposited in Qinghai-Tibetan Plateau Museum of Biology (HNWP), Northwest Institute of Plateau Biology, Chinese Academy of Sciences. The chloroplast DNA library was prepared by following protocol (Chi et al. Citation2018; Thomson et al. Citation2018) with a fragment size of 150 pb. The sequencing was performed using NovaSeq 6000 (Illumina Inc., San Diego, CA, USA) with a paired-end (PE) approach. After filtering and assembling of the raw reads, the contigs are aligned to the reference plastome. The assembled chloroplast genome has annotated using Plann (Huang and Cronk Citation2015) and the error in annotation has corrected using Geneious (Matthew et al. Citation2012). The complete cpDNA sequence of C. falcatum has submitted to GenBank (Accession number MK331815).
The complete genome size of C. falcatum is 151,423 bp in length, containing the large single copy (LSC, 81,721 bp), a small single copy (SSC, 18,248 bp), and two inverted repeats (IR, 25,727 pb) regions. The overall GC contents of this chloroplast genomes are 38.26%. In total, 133 unique genes are annotated, including 88 protein-coding genes, 8 rRNA genes, and 37 tRNA genes. The protein-coding genes, rRNA genes, and tRNA genes account for 66.2, 6.0, and 27.8% of all annotated genes, respectively.
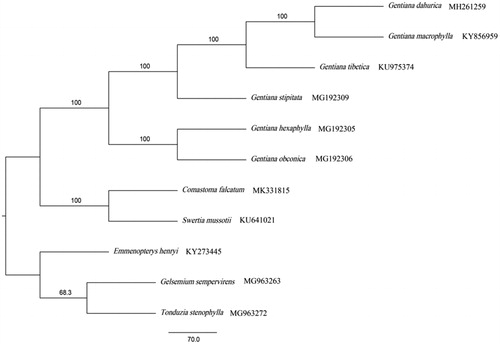
The maximum parsimony phylogenetic tree was generated based on the complete chloroplast genome of C. falcatum and other species which have a close genetic relationship (). The results showed that C. falcatum is closely related to Swertia mussotii. This study of complete chloroplast genome sequencing of C. falcatum will provide useful information for future studies to solve the problems related to the phylogeny of genus Comastoma.
Figure 1. Maximum parsimony phylogenetic tree of C. falcatum and other 10 species with Emmenopterys henryi, Gelsemium sempervirens, and Tonduzia stenophylla as outgroups from the Gentianales. This tree is based on complete chloroplast genome sequences. Bootstrap support values are displayed on each node.
Disclosure statement
The authors declare no conflicts of interest and are responsible for the content.
Additional information
Funding
References
- Brakefield PM. 2006. Evo-devo and constraints on selection. Trends Ecol Evol (Amst.). 21:362–368.
- Chassot P, Nemomissa S, Yuan YM, Küpfer P. 2001. High paraphyly of Swertia L. (Gentianaceae) in the Gentianella-lineage as revealed by nuclear and chloroplast DNA sequence variation. Plant Syst Evol. 229:1–21.
- Chi X, Wang J, Gao Q, Zhang F, Chen S. 2018. The complete chloroplast genomes of two Lancea species with comparative analysis. Molecules. 23:602.
- Doyle JJ, Doyle JL. 1987. A Rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem Bull. 19:11–15.
- Hagen KBV, Kadereit JW. 2001. The phylogeny of Gentianella (Gentianaceae) and its colonization of the southern hemisphere as revealed by nuclear and chloroplast DNA sequence variation. Org Divers Evol. 1:61–79.
- Hagen KBV, Kadereit JW. 2002. Phylogeny and flower evolution of the Swertiinae (Gentianaceae-Gentianeae): homoplasy and the principle of variable proportions. Syst Bot. 27:548–572.
- Ho T, James SP. 1995.Comastoma. In: Wu Z, Raven PH, editors. Flora of China. Beijing: Science Press and Missouri Botanical Garden; p. 1–139.
- Huang DI, Cronk QC. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026.
- Matthew K, Richard M, Amy W, Steven SH, Matthew C, Shane S, Simon B, Alex C, Sidney M, Chris D. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647.
- Thomson AM, Vargas OM, Dick CW. 2018. Complete plastome sequences from Bertholletia excelsa and 23 related species yield informative markers for Lecythidaceae. Appl Plant Sci. 6:e01151.
- Yuan Y-M, Küpfer P. 1995. Molecular phylogenetics of the subtribe Gentianinae (Gentianaceae) inferred from the sequences of internal transcribed spacers (ITS) of nuclear ribosomal DNA. Plant Syst Evol. 196:207–226.
