Abstract
Pyrus ussuriensis Maxim. is an oriental domesticated pear which have been used as a traditional medicine as well as dominant contributor of many Asian origin pear breeds. Here, we presented complete chloroplast genome of P. ussuriensis which is 160,157 bp long and has four subregions: 88,148 bp of large single-copy (LSC) and 19,227 bp of small singl- copy (SSC) regions are separated by 26,391 bp of inverted repeat (IR) regions including 130 genes (85 protein-coding genes, 8 rRNAs, and 37 tRNAs). The overall GC content of the chloroplast genome is 36.6% and in the LSC, SSC, and IR regions are 34.2%, 30.4%, and 42.7%, respectively. Phylogenetic trees reflect that P. ussuriensis has been utilized as a dominant contributor to many Asian origin pear breeds.
The genus Pyrus L. (tribe Maleae, Rosaceae) includes ∼25 species and mainly occurs in the northern part of Africa, Europe, and Asia (Rubtsov Citation1944; Wu et al. Citation2003). It has been geographically divided into two groups: occidental pears in western Eurasia and oriental pears in eastern Asia. Both were domesticated as important food resources in these two distinct regions (Rubtsov Citation1944). However, phylogenetic relationship of this genus is poorly resolved partly due to rapid radiation and extensive inter-specific hybridization (Zheng et al. Citation2014). Pyrus ussuriensis Maxim., one of the oriental species, is distributed in China, Russia, Korea, and Japan. It has edible fruits, which are also used as traditional medicine (Kim and Song Citation2012). This species is also known as the dominant contributor for many Asian origin pear breeds (Wu et al. Citation2003; Cao et al. Citation2012).
Total DNA of P. ussuriensis collected in Mt. Hambaek, Gangwon Province, Korea (Voucher in InfoBoss Cyber Herbarium (IN); IB-00586), which was extracted from fresh leaves using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Genome sequencing was performed using HiSeqX at Macrogen Inc., Korea, and de novo assembly and confirmation were done by Velvet 1.2.10 (Zerbino and Birney Citation2008), SOAPGapCloser 1.12 (Zhao et al. Citation2011), BWA 0.7.17 (Li Citation2013), and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.0.5 (Biomatters Ltd, Auckland, New Zealand) was used for chloroplast genome annotation based on Pyrus pyrifolia chloroplast genome (NC_015996; Terakami et al. Citation2012)
The chloroplast genome of P. ussuriensis (Genbank accession is MK172841) is 160,157 bp and has four subregions: 88,148 bp of large single-copy (LSC) and 19,227 bp of small single-copy (SSC) regions are separated by 26,391 bp of the inverted repeat (IR). It contains 130 genes (85 protein-coding genes, 8 rRNAs, and 37 tRNAs); among them 19 genes (8 protein-coding genes, 4 rRNAs, and 7 tRNAs) are duplicated in IR regions. The overall GC content of P. ussuriensis is 36.6% and those in the LSC, SSC, and IR regions are 34.2%, 30.4%, and 42.7%, respectively.
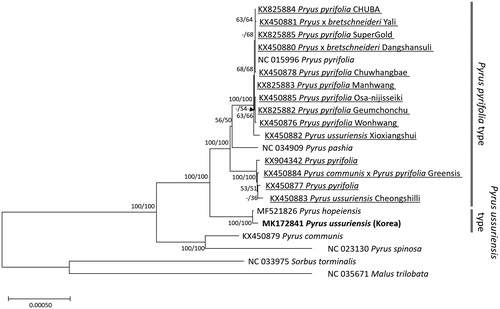
Twenty Pyrus including 14 domesticated pears and 2 chloroplast genomes in Rosaceae as an outgroup were used for constructing phylogenic trees. Whole chloroplast genome sequences were aligned by MAFFT 7.388 (Katoh and Standley Citation2013) for constructing neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) trees using MEGA X (Kumar et al. Citation2018). Phylogenetic trees show that three P. ussuriensis chloroplast genomes (MK172841, KX450883, and KX450882) are scattered in different clades (). Insertion between accD and psaI (54,638–54,725 bp), which can distinguish P. ussuriensis type (Type A) and P. pyrifolia type (Type B), confirms that our chloroplast genome is P. ussuriensis type (Type A); while the rest two genomes are P. pyrifolia type (Type B; Zheng et al. Citation2014). In addition, P. pyrifolia chloroplast genomes are separated into two clades, corresponding to B5 and B6 clades in Zheng et al. (Citation2014) (), reflecting that P. ussuriensis has been utilized as a dominant contributor of many Asian origin pear breeds.
Figure 1 Neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1000) phylogenetic trees of 20 Pyrus and two outgroup complete chloroplast genomes: 3 Pyrus ussuriensis (MK172841, in this study, KX450883 and KX450882; unpublished), 10 Pyrus pyrifolia (KX825884, KX825885, NC 015996, KX450878, KX825883, KX450885, KX825882, KX450876, KX904342, and KX450877), 2 Pryus x bretschneideri (KX450881 and KX450880), Pyrus pashia (NC_034909), Pyrus communis x Pyrus pyrifolia (KX450884), Pyrus hopensis (MF521826), Pyrus communis (KX450879), Pyrus spinosa (NC 023130), Sorbus torminalis (NC 033975), and Malus trilobata (NC_035671). Underlines in species name indicate that those sequences are from domesticated pears. Grey bars indicate two major clades of Pyrus: Pyrus pyrifolia type is corresponding to B3 to B6 clades and Pyrus ussuriensis type is to B1 and B2 clades (Zheng et al.Citation2014). Phylogenetic tree was displayed based on neighbor joining tree. The numbers above branches indicate bootstrap support values of maximum likelihood and neighbor joining phylogenetic tree, respectively.
Disclosure statement
The authors declare that they have no competing interests.
Additional information
Funding
References
- Cao Y, Tian L, Gao Y, Liu F. 2012. Genetic diversity of cultivated and wild Ussurian Pear (Pyrus ussuriensis Maxim.) in China evaluated with M13-tailed SSR markers. Genetic Res Crop Evol. 59:9–17.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kim H, Song M-J. 2012. Traditional plant-based therapies for respiratory diseases found in North Jeolla Province, Korea. J Altern Complement Med. 18:287–293.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. ScienceOpen. arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Rubtsov G. 1944. Geographical distribution of the genus Pyrus and trends and factors in its evolution. The Am Naturalist. 78:358–366.
- Terakami S, Matsumura Y, Kurita K, Kanamori H, Katayose Y, Yamamoto T, Katayama H. 2012. Complete sequence of the chloroplast genome from pear (Pyrus pyrifolia): genome structure and comparative analysis. Tree Genet Genom. 8:841–854.
- Wu Z, Raven P, Hong D. 2003. Flora of China. Vol. 9, Pittosporaceae through Connaraceae. Beijing (China): Science Press.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genom Res. 18:821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12:S2.
- Zheng X, Cai D, Potter D, Postman J, Liu J, Teng Y. 2014. Phylogeny and evolutionary histories of Pyrus L. revealed by phylogenetic trees and networks based on data from multiple DNA sequences. Mol Phylogenet Evol. 80:54–65.
