Abstract
Complete mitochondrial genome of Onustus exutus has been sequenced and phylogenetic relationships were evaluated. The circular mitochondrial genome is 16,043 bp in length consisting of 13 protein-coding genes, 22 transfer RNA genes, and 2 ribosomal RNA genes. The nucleotide composition of the complete mitochondrial genome of O. exutus is 30.46% A, 14.32% C, 15.83% G, and 39.38% T. With the exception of eight tRNA genes, all other mitochondrial genes are encoded on the heavy strand. Thirteen protein-coding genes start with ATG. For the stop codon, CYTB, ND6, ND4L, and ND4 end with TAG, other 9 genes end with TAA. Phylogenetic tree was constructed based on 13 protein-coding gene sequences of 11 species using the neighbor-joining method, and the result showed that O. exutus is most closely related to Conomurex luhuanus. This is the first record of complete mitochondrial genome from the genus in the Onustus. The present study provides additional data for Mesogastropoda phylogeny.
The Onustus is commonly called carrier shells, which is a genus of medium-sized to large sea snails (Kreipl Citation1999). Onustus exutus (Reeve, 1842) belongs to the group Gastropoda, family Xenophoridae, genus Onustus. It is mainly distributed from India to the western Pacific Ocean. It is an unusual species found in the South China Sea, which is a carnivorous snail with a low conical shape, usually lives on a sandy seabed 20–40 m deep (Gu Citation2005). The shell is thin and irregular, it can reach about 7 cm in length. Information on the genetic of O. exutus is limited. It is the first species in the family Xenophoridae for which complete mitochondrial genome has been determined in this study. In order to obtain genetic information and understand the evolution of O. exutus, we sequenced its complete mitochondrial genome (GenBank accession no. MK327366).
The sample of Onustus exutus were collected from the sandy bottom of Zhoushan, Zhejiang province, China (122°6E, 30°5N), in July 2018. The specimen was identified based on the morphologic features by experienced fisheries researchers. Muscle tissues of individual specimens for molecular analysis were reserved in ethanol absolute. The whole genomic DNA of O. exutus was extracted using the phenol–chloroform method (Barnett and Larson Citation2012). The quantity and quality of the isolated DNA was examined by NanoDrop 2000 spectrophotometer (Thermo Scientific) and by electrophoresis in 1% agarose gel, and then diluted to a final concentration of 50–60 ng/µl in 1 × TE buffer and stored at 4 °C for further analysis. The complete mitochondrial genome of O. exutus was sequenced by Illumina Hiseq2500 platform using the total genomic DNA in Yuanxin Biomedical Technology Co., Ltd Shanghai, China.
The complete mitochondrial genome of O. exutus was 16,043 bp in length, consisting of 13 protein-coding genes, 2 ribosomal RNA genes (including 12S rRNA and 16S rRNA), and 22 transfer RNA genes (tRNA). The nucleotide base composition is A (30.46%), T (39.38%), C (14.32%) and G (15.83%), respectively. The A + T content (69.84%) is higher than G + C content (30.16%). Except eight tRNAs (Met, Tyr, Cys, Trp, Leu, Gly, Glu, Thr), other genes were encoded on the heavy strand. All 13 protein-coding genes start with ATG. For the stop codon, nine protein-coding genes (COX1, COX2, COX3, ND1, ND2, ND3, ND5, ATP6, and ATP8) end with TAA, and four genes (CYTB, ND6, ND4L and ND4) end with TAG. The two ribosomal RNA genesare allocated as follows: 16S rRNA (1348 bp) is located between tRNAVal and tRNALeu genes and 12S rRNA (886 bp) is located between tRNAGlu and tRNAVal genes.
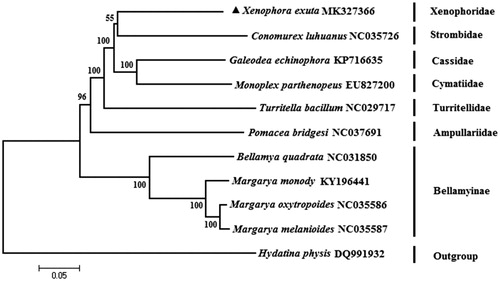
The phylogenetic tree () was constructed using the neighbor-joining method (Saitou and Nei Citation1987) based on the 13 protein-coding genes, with a Hydatinidae species as the outgroup. The result shows that O. exutus was most closely related to C. luhuanus. Earlier reported morphology-based phylogenetic (Simone Citation2005) and behavioral (Berg Citation1975) study of Strombidae and Xenophoridae showed similar results. We expect the present results will further facilitate for the study on the analysis of the evolution of Mesogastropoda.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Barnett R, Larson G. 2012. A phenol-chloroform protocol for extracting DNA from ancient samples. Methods Mol Biol. 840:13–19.
- Berg CJ. 1975. Behavior and ecology of conch (Superfamily Strombacea) on a deep subtidal algal plain. Bull Mar Sci. 25:307–317.
- Gu M. 2005. Atlas of the ell in Famous Shell in the World for identification and Appreciation. Zhengzhou city, Henan province, China: Henan Scientific and Technological Publishing House.
- Kreipl K, AA. 1999. Recent Xenophoridae. Hackenheim (Germany): Conchbooks.
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4:406–425.
- Simone LRL. 2005. Comparative morphological study of representatives of the three families of Stromboidea and the Xenophoroidea (Mollusca, Caenogastropoda), with an assessment of their phylogeny. Arq Zool. 37:141.