Abstract
We describe the complete mitochondrial genome of the broadclub cuttlefish Sepia latimanus, the second largest cuttlefish, collected from the coral waters of Naozhou Island in tropical South China. It is a circular molecule of 16,225 bp in size and consists of 13 protein-coding genes, 2 rRNA genes, 22 tRNA genes, and 1 control region in typical gene arrangement conforming to the vertebrate consensus. The complete mitochondrial genomic sequence of S. latimanus and other 8 species from the same Family Sepiidae were used for phylogenetic analysis and demonstrated that S. latimanus clustered with S. apama, the largest cuttlefish species, and made a separate clade, closely related to the other Sepia and Sepiella species in Family Sepiidae.
Sepia latimanus is widely distributed in tropical and warm temperate waters across the Indian and western Pacific Oceans (Dan et al. Citation2012). It is the second biggest cuttlefish after S. apama with a maximum dorsal mantle length of 50 cm and a total weight of 10 kg and become important for fisheries in many countries (Pratasik et al. Citation2015). In China, S.latimanus supports local fisheries in Guangdong province, and the annual production is hundreds of tons in history (Dong Citation1988). However, there is little genetic information about this species available in China. Here, we first sequenced and characterized the mitogenome of S. latimanus in China and examined its phylogenetic position within Sepiidae.
The samples used in this study were collected from coral reef waters in Naozhou sea area(110°37″E; 20°50 N)during spring, 2016. After sampling, the specimens were stored in 95% ethanol and transported to the university’s laboratory. Total genomic DNA was extracted from the muscle by proteinase K digestion, following a standard phenol–chloroform method (Sambrook et al. Citation1989). Twenty sets of primers were designed to amplify the complete mitochondrial genome sequences. The annotation of mitochondrial protein-coding genes and ribosomal RNA genes were identified using DOGMA (Wyman et al. Citation2004). MITOS was used to predict the transfer RNA genes (Bernt et al. Citation2013). The mitogenome has been deposited in the GeneBank database (accession number: MK347498).
The mitogenome sequence is 16,225bp in length, containing 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and a non-coding region. The overall nucleotide composition is 41.1% A, 35.8% T, 14.9% C, 8.1% G, with AT bias of 76.9% (Shadel and Clayton Citation1997). Twenty-two of the genes are encoded on the light strand, while the other 15 on the heavy strand.
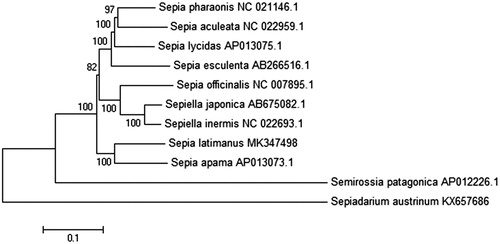
Nine mitogenome sequences from Sepiidae species including the one obtained in this study were used for phylogenetic analysis. Sepiadarium austrinum and Semirossia patagonica in the same order were used as an outgroup. Phylogenetic reconstruction was performed using neighbor-joining (NJ) with Meg 5.0 (Tamura et al. Citation2011) with a bootstrap of 1000 replicates (). Results showed that S. latimanus was most closely related to S. apama, the largest cuttlefish species, and made a separate branch in Family Sepiidae. The rest of species, five from genus Sepia and two from genus Sepiella, are more closely related and made another separate branch. Similar results were obtained by Yoshida et al. (Citation2006), who found Sepiidae are polyphyletic and can be branched into four clades based on three mitochondrial gene sequences, and S.latimanus was closely related to Metasepia tullbergi and made a separate clade, and the other three clades are made of species from genus Sepia and Sepiella that cannot be separated by phylogenetic analysis. The reason for this unexpected confusion about genus Sepia and Sepiella may come from the unsolved classification in Sepiidae species (Yoshida et al. Citation2006). However, the results of this study will provide valuable genetic information for S. latimanus in South China sea and accumulate useful data for further evolutionary analysis of Sepiidae species.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Dan S, Hamasaki K, Yamashita T, et al. 2012. Age-based life cycle traits of the broadclub cuttlefish Sepia latimanus confirmed through release–recapture experiments. Aquat Biol. 17:181–195.
- Dong ZZ. 1988. Fauna sinica: phylum mollusca (Class Cephalopoda) (in Chinese). Beijing (China): Science Press.
- Pratasik SB, Arfiati MD, Setyohadi D. 2015. Size at first maturity of cuttlefish, Sepia latimanus, from North Sulawesi Waters, Indonesia. Mar Sci. 5:6–10.
- Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press.
- Shadel GS, Clayton DA. 1997. Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem. 66:409–435.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yoshida M, Tsuneki K, Furuya H. 2006. Phylogeny of selected Sepiidae (Mollusca, Cephalopoda) based on 12S, 16S, and COI sequences, with comments on the taxonomic reliability of several morphological characters. Zool Sci. 23: 341–351.