Abstract
Holothuria (Stauropora) pervicax was a common, widespread tropical holothuroid. The complete mitogenome of H. (S.) pervicax was 15,790 bp in length and consisted of 13 PCGs, 22 tRNAs, and two rRNAs. The gene order and genetic characteristics were same with other mitogenomes of Holothuria. The phylogenetic tree revealed that clade of Holothuria was clearly monophyletic, included H. (S.) pervicax.
Holothuria (Stauropora) pervicax Selenka, 1867, which belongs to the family Holothuriidae Burmeister, 1837, was a common, widespread tropical holothuroid, and occurs in Korea, southern Japan, and Hawaii (Clark and Rowe Citation1971; Rowe and Gates Citation1995; Shin Citation2012). In this study, we presented a complete mitogenome of H. (S.) pervicax from Jeju Island, Korea.
Specimens of H. (S.) pervicax were collected by SCUBA diving at a depth of 21 m in adjacent waters of Jeju Island, Korea (33°13′39.7″N, 126°34′02.8″E). Voucher specimens and mitochondrial DNA sample were deposited in the Marine Echinoderm Resources Bank of Korea (Seoul, Korea) and granted a voucher number. Mitochondrial DNA (mtDNA) extraction, amplification, and sequencing were performed according to Lee and Shin (Citation2018). The complete mitogenome was annotated using Geneious ver. 11.1.5 (Biomatters Ltd, Auckland, New Zealand). Transfer RNA (tRNA) genes were identified using tRNAscan-SE online (Lowe and Chan Citation2016), with the following search mode: sequence source with ‘other mitochondrial’ selected, and genetic code for tRNA isotype prediction with “Echinoderm Mito” selected. The gene locations were determined using DOGMA (Wyman et al. Citation2004). A phylogenetic analysis was performed with nine complete mitogenomes of holothuroids, including H. (S.) pervicax, and two echinoids, Mesocentrotus franciscanus (NC_024177) and M. nudus (NC_020771) were used as outgroups for this analysis. Mitogenome sequences were aligned using MAFFT (Katoh and Standley Citation2013). The mitogenome dataset was analyzed using maximum likelihood (ML) with RAxML 8.2 (Stamatakis Citation2014) and Bayesian inference (BI) with MrBayes 3.2 (Ronquist and Huelsenbeck Citation2003). The best-fit substitution was estimated using jModelTest 2.1.1 (Guindon and Gascuel Citation2003; Darriba et al. Citation2012) for the nucleotide dataset of 13 protein-coding genes (PCGs) and two rRNAs, respectively. For ML analyses, bootstrap resampling was performed using the rapid option with 1000 iterations. For BI analyses, the Markov chain Monte Carlo (MCMC) was run for 10,000,000 generations, with sampling every 1000 generations.
The complete mitogenome of H. (S.) pervicax (GenBank accession No. MK328500) was 15,790 bp in length and contained 13 PCGs, 22 tRNA genes, and two rRNA genes. The gene order and direction of PCGs and rRNAs were identical to those of the other eight complete mitogenomes of holothuroids species. All of the PCGs contained an ATG initiation codon (Methionine). The termination codons of the PCGs were TAA and TAG codons without the cytochrome oxidase subunit II, glutamic acid (GAA)+T.
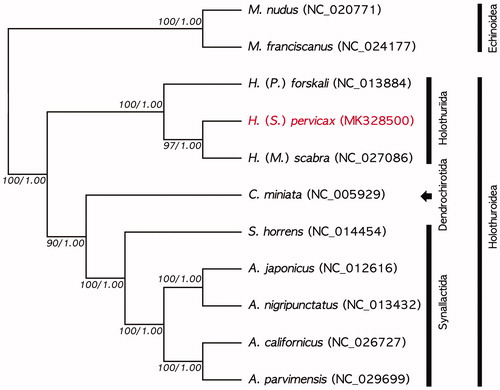
In the phylogenetic tree, H. (S.) pervicax was grouped distinctly with the other species in the genus Holothuria, H. (Panningothuria) forskali (NC_013884) and H. (Metriatyla) scabra (NC_027086) (). The Holothuria clade was monophyletic and was clearly separated from other taxa, order Dendrochirotida and synallactida. As the result, this mitogenome will be a resource for further phylogenetic studies of mitogenome in holothuroids.
Figure 1. Phylogenetic tree of maximum likelihood (ML) and Bayesian inference (BI) method based on the nucleotide sequences of 13 PCGs and two rRNAs of nine holothuroids, included H. (S.) pervicax (MK328500), and two echinoids. Bootstrap support and posterior properties values were indicated on each node as >70 and 0.7.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Clark AM, Rowe FWE. 1971. Monograph of shallow-water Indo-West Pacific echinoderms. Trustees of the British Museum of Natural History. London. 1–238.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772.
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biol. 52:696–704.
- Katoh K, Standley DM. 2013. MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Lee T, Shin S. 2018. Complete mitochondrial genome analysis of Distolasterias nipon (Echinodermata, Asteroidea, Forcipulatida). Mitochondr DNA B. 3:1290–1291.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44:W54–W57.
- Ronquist F, Huelsenbeck JP. 2003. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Rowe FWE, Gates J. 1995. Echinodermata. In: A. Wells, editor. Zoological catalogue of Australia. Vol. 33. Melbourne: CSIRO Australia; p. 1–510.
- Shin S. 2012. Sea Cucumbers: Echinodermata: Echinozoa: Holothuroidea. Invertebrate Fauna of Korea. 4th ed. Vol. 32. Incheon: National Institute of Biological Resources; p. 1–127.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
