Abstract
The complete mitochondrial genome was sequenced from the KIT strain of the freshwater water flea (Daphnia magna). The sequenced genome size was 15,253 bp long, possessing identical gene order and contents to those of the congeneric species Daphnia galeata in the genus Daphnia, while the direction of ND2 gene in some strains of D. magna was opposite. The mitochondrial genome of D. magna has 13 protein-coding genes (PGCs), 2 rRNAs, and 22 tRNAs. Of 13 PGCs, five genes (ND2, CO1, CO2, ND4, and ND5) had incomplete stop codons. Furthermore, the stop codons of the remaining eight PGCs were TAG and TAA, while the start codon of CO1 gene was CTA. The base composition of D. magna mitogenome shows an anti-G and -C bias (17.1% and 17.2%) on 13 PCGs, respectively.
Keywords:
To date, more than 200 species are retrieved in the genus Daphnia (Kotov et al. Citation2012, http://fada.biodiversity.be/CheckLists/Crustacea-Cladocera.pdf). Among them, the complete mitochondrial genome of Daphnia pulex has been firstly reported (Crease Citation1999) with high mutation rates of mitochondrial genome (Xu et al. Citation2012). As one of the promising model organisms in the freshwater environment, Daphnia magna has been widely used in genetics, developmental biology, environmental toxicology, and other aspects. In this paper, we report the complete mitochondrial genome of the freshwater water flea D. magna to better understand the phylogenetic position of and within the genus Daphnia.
The water flea D. magna (KIT strain) was originally purchased from CarolineTM Science and Math (Burlington, NC) by Korean Institute of Toxicology (KIT) (Daejeon, South Korea) on 5 May 2006 and maintained in the Department of Biological Science, Sungkyunkwan University in South Korea (kindly provided by Prof. Jinhee Choi, University of Seoul in South Korea). We sequenced the whole genome of the water flea D. magna from whole body genomic DNA with several paired-end and mate-pair libraries using the Illumina HiSeq 2500 platform (GenomeAnalyzer, Illumina, San Diego, CA). De novo assembly was conducted by CLC Assembly Cell (https://www.qiagenbioinformatics.com/). Of the assembled 264,740 D. magna contigs, a single contig was mapped to the mitochondrial DNA of D. galeata (GenBank accession no. NC_034297).
The total length of the complete mitochondrial genome of D. magna is 15,253 bp (GenBank accession no. MK370029). The mitochondrial genome of D. magna contains 13 protein-coding genes (PGCs), 2 rRNAs, and 22 tRNAs. The direction of 13 PGCs was identical to those of a congeneric species D. galeata (Tokishita et al. Citation2017). Between two sister species (D. magna and D. galeata), the similarities of amino acids and nucleotides of 13 PCGs were 73.7% and 67.88%, respectively. Of 13 PCGs in D. magna, five genes (ND2, CO1, CO2, ND5, and ND4) had incomplete stop codons. The mitochondrial genome base composition of 13 PCGs was 25.4% for A, 40.3% for T, 17.1% for G, and 17.2% for C. The A + T base composition (65.7%) was slightly higher than G + C (34.3%). Particularly, in D. magna KIT strain, there is an anti-G bias (14.7% and 14.2%) at the second and third position of codons and an anti-C bias (18.1% and 14.9%) at the second and third position of codons. The most commonly used start codon was CTA, ATC, and ATT for CO1, ND3, and ND6 genes, respectively, in the freshwater water flea D. magna.
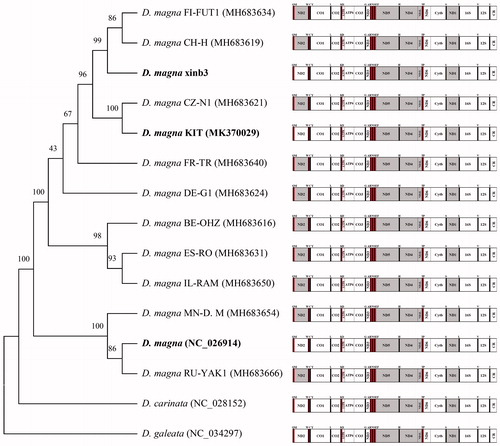
The placement of D. magna KIT strain among 13 D. magna strains with known complete mitogenomes with outgroup (D. galeata) is shown in . In 15 daphnids, all the components (PGCs, rRNAs, tRNAs) were identically placed. However, the direction of ND2 gene of three strains (xinb3, KIT, and GenBank no. NC_026914) of D. magna, was forward as shown in other two congeneric species (D. carinata and D. galeata), while others demonstrated the opposite. D. magna KIT strain clustered closely to D. magna CZ–N1 strain.
Figure 1. Phylogenetic analysis. We conducted a comparison of the mitochondrial genomes of 15 daphnids. Twelve protein-coding genes of each mitochondrial genome except for ND2 gene were aligned using MEGA software (ver. 7.0) with the ClustalW alignment algorithm. To establish the best-fit substitution model for phylogenetic analysis, the model with the lowest Bayesian Information Criterion (BIC) and Akaike Information Criterion (AIC) scores were estimated using a maximum likelihood (ML) analysis. According to the results of model test, maximum likelihood phylogenetic analyses were performed with the LG + G + I model. Gray, the opposite direction of PCGs.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Crease TJ. 1999. The complete sequence of the mitochondrial genome of Daphnia pulex (Cladocera: Crustacea). Gene. 233:89–99.
- Kotov A, Forró L, Korovchinsky NM, Petrusek A. 2012. Crustacea-Cladocera checklist. World checklist of freshwater Cladocera species. Belgian Biodiversity Platform. Retrieved October 29, 2012.
- Tokishita SI, Shibuya H, Kobayashi T, Sakamoto M, Ha JY, Yokobori SI, Yamagata H, Hanazato T. 2017. Diversification of mitochondrial genome of Daphnia galeata (Cladocera, Crustacea): comparison with phylogenetic consideration of the complete sequences of clones isolated from five lakes in Japan. Gene. 611:38–46.
- Xu S, Schaack S, Seyfert A, Choi E, Lynch M, Cristescu ME. 2012. High mutation rates in the mitochondrial genomes of Daphnia pulex. Mol Biol Evol. 29:763–769.
