Abstract
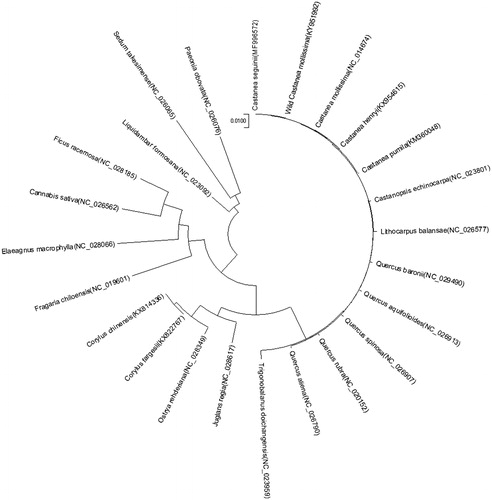
The complete chloroplast genome sequence of an endemic wild Chinese tree Castanea seguinii was determined. The complete chloroplast genome is 160,862 bp and contains a pair of inverted repeat regions of 25,720 bp each, a large single-copy region of 90,440 bp, and a small single-copy region of 18,982 bp. The genome harbors 113 genes, including 79 protein-coding genes, four ribosomal RNA genes, and 30 transfer RNA genes. A phylogenetic analysis based on chloroplast genomes indicates that wild C. seguinii is closely related to wild Castanea mollissim.
Castanea seguinii, an endemic Chinese species of Fagaceae castanea, which are shrubs or small trees (Rutter et al. Citation1990), is widely distributed in Shandong, Henan, Shanxi, Shaanxi and other provinces south of the Yangtze River Basin. The species has strong germination and early fruiting characteristics (annual seedlings or sprouts can blossom and bear fruit). Although several chloroplast (cp) DNA markers have previously been reported(Lang et al. Citation2006; Cheng et al. Citation2012; Yousefzadeh et al. Citation2014), little is known about the cp genome of C. seguinii. In the present study, we report the first complete cp genome of C. seguinii based on Illumina paired-end sequencing data (GenBank accession number MF996572).
Fresh leaves were collected from a single wild C. seguinii tree growing on Dabie Mountain in Jinzhai County (115°778′312″N, 31°134′837″E), Anhui, China. Voucher specimens were deposited at Beijing Academy of Forestry and Pomology Sciences Herbarium. DNA extraction was performed according to a modified CTAB protocol (Li et al. Citation2013). High-throughput sequencing was carried out using the HiSeq4000 PE150 System (Illumina, San Diego, CA, USA). A total of 3,019,700 500-bp raw reads were obtained and used for the cp genome assembly with SPAdes 3.6.1 (Bankevich et al. Citation2012) and SOAPdenovo software (Luo et al. Citation2012). A reference-guided assembly was then performed to reconstruct the cp genome with the BLAST program (Altschul et al. Citation1990) using closely related species as references. After filling the gaps with GapCloser (http://soap.genomics.org.cn/index.html), a 160,869-bp cp genome was obtained for C. seguinii. The annotation was performed using the Dual Organellar GenoMe Annotator to generate a physical map of the cp genome (Wyman et al. Citation2004).
The circular cp genome of C. seguinii contains a pair of inverted repeat regions of 25,720 bp each, and large and small single-copy regions of 90,440 bp and 18,982 bp, respectively. The genome comprises 113 genes, including 79 protein-coding, four ribosomal RNA, and 30 transfer RNA genes. Interestingly, there is a trans-splicing gene (rps12) whose 5′ exon is located in the LSC, whereas the 3′ exon is located in the IR.
To identify the phylogenetic position of C. seguinii, a maximum likelihood analysis was performed based on 13 plant cp genomes from the order Fagales and 11 other outgroup plants using MEGA6 software (Tamura et al. Citation2013). The cp genome of C. seguinii was closely related to that of wild Castanea mollissim (). The new information provided by the cp genome will be important for formulating new conservation and management strategies for wild C. seguinii.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Cheng L, Feng H, Rao Q, Wu W, Zhou M, Hu G, Huang W. 2012. Diversity of wild chestnut chloroplast DNA SSRs in Shiyan. J Fruit Sci Chinese. 29:382–386.
- Lang P, Dane F, Kubisiak TL. 2006. Phylogeny of Castanea (Fagaceae) based on chloroplast trnT-L-F sequence data. Tree Genet Genom. 2:132–139.
- Li J, Wang S, Jing Y, Wang L, Zhou S. 2013. A modified CTAB protocol for plant DNA extraction. Chinese Bull Bot. 48:72–78. Chinese.
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 1:18.
- Rutter P, Miller G, Payne J. 1990. Chestnuts In: Moore JN, Ballington JR, editors. Genetic resources of temperate fruit and nut crops. Wageningen (The Netherlands): The International Society for Horticultural Science; p. 761–788.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 30:2725–2729.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yousefzadeh H, Hosseinzadeh Colagar A, Akbarzadeh F, Tippery NP. 2014. Taxonomic status and genetic differentiation of Hyrcanian Castanea based on noncoding chloroplast DNA sequences data. Tree Genetics & Genomes. 10:1611–1629.