Abstract
Herein, we first determined the complete mitochondrial genome of Turricula nelliae spurius in this study. The circular mitochondrial genome is 16,450 bp in length, consisting of 13 protein-coding genes, 22 tRNA genes, and two ribosomal RNA genes. The nucleotide composition is 15.0% of C, 15.8% of G, 38.0% of T, and 31.2% of A. The control region is between tRNAPhe and COX3 genes and has 1,144 bp. In 13 protein-coding genes, three genes (nad2, nad4, atp6) start with ATA, the other 10 genes are with ATG. For the stop codon, nad3 and nad4l end with TAG, other 11 genes end with TAA. The phylogenetic tree was constructed based on 13 protein-coding genes of T. nelliae spurius and other 11 species using the Neighbor-joining method, and the result showed that T. nelliae spurius is closed to the Gemmuloborsonia moosai and clustered in the same branch with its same family species—Lophiotoma cerithiformis. The mitochondrial genome would be helpful for the study of population genetic and phylogenetic analysis of the family Turridae.
Turricula nelliae spurius (Gastropoda, Turridae) is one of the carnivores sea snail (Bouchet et al., Citation2011). The shell size of adult varies between 25 and 40 mm, the turreted shell has a fusiform shape, and it contains 14 strongly excavated whorls (Smith Citation1877). It is a common species that can be found in the subtidal zone. It is widely distributed in the Indo-Pacific region. It is usually used as the ornaments in the past by its unique appearance. At present, only a few preliminary taxonomic and life cycle studies on T. nelliae spurius (Miller and Morton Citation1990), and the study of genetic or phylogenetic is still blank.
In this study, it is the first study that determined the complete mitochondrial genome of T. nelliae spurius. The specimen of T. nelliae spurius was collected in Zhejiang province, China (122.2°E, 30.3°N) and identified by morphology and deposited in Zhejiang Ocean University. The total DNA extraction was utilized the salting-out method (Aljanabi and Martinez Citation1997) with the muscle. Then, total genomic DNA was diluted to a final concentration of 60–80 ng/μl in 1 × TE buffer and stored at 4 °C in our laboratory at Zhejiang Ocean University for further analysis. The genomic DNA was prepared in 350 bp paired-end libraries. The Illumina HiSeq X Ten platform was used to perform the high-throughput sequence.
The complete mitochondrial genome of T. nelliae spurius is 16,450 bp in length (GenBank accession number: MK251986). The complete mitochondrial genome has two ribosomal RNA genes, 22 transfer RNA (tRNA) genes, and 13 protein-coding genes. The control region is between tRNAPhe and COX3 genes and has 1144 bp. The nucleotide composition for T. nelliae spurius is 15.0% of C, 15.8% of G, 38.0% of T, and 31.2% of A. In 13 protein-coding genes, three genes (nad2, nad4, atp6) start with ATA, the other ten genes are with ATG. For the stop codon, nad3 and nad4l end with TAG, other 11 genes end with TAA. The 12S rRNA is 882 bp between the tRNAGlu and tRNAVal, and the 16S rRNA is 1353 bp between the tRNAVal and tRNALeu1. The tRNAMet, Tyr, Cys, Trp, Gln, Gly, Glu, Thr are translated by the light chain.
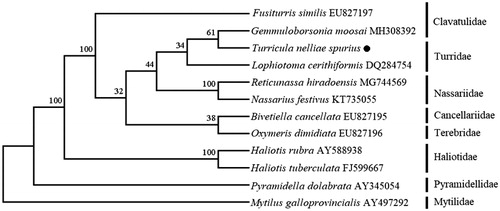
The phylogenetic tree () was constructed based on 13 protein-coding genes of the T. nelliae spurius and other 11 species using the Neighbor-joining method (Saitou and Nei Citation1987) in the program Phylip (Felsenstein Citation1989). The tree showed that the T. nelliae spurius is closed to the Gemmuloborsonia moosai (MH308392) and clustered in the same branch with its same family species—Lophiotoma cerithiformis (DQ284754). We believe that this result will further supplement the genome information in mitochondria of the family Turridae and facilitate the study on population genetic.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Aljanabi SM, Martinez I. 1997. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 25:4692–4693.
- Bouchet P, Kantor YI, Sysoev A, Puillandre N. 2011. A new operational classification of the Conoidea (Gastropoda). J Molluscan Stud. 77:273–308.
- Felsenstein J. 1989. PHYLIP-Phylogeny inference package (Version 3.2). Cladistics. 5:164–166.
- Miller JA, Morton B. 1990. The feeding and prey capture mechanism of Turricula nelliae spurius (Hedley) (Gastropoda: Turridae). Mar Flora Fauna Hong Kong South China II Behav Morphol Physiol Pollut. 3:979.
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4:406–425.
- Smith EA. 1877. On the shells of Lake Nyasa, and on a few marine species from Mozambique. Proc Zoo Soc London. 1877: 712–722.