Abstract
The complete mitochondrial genome of the Epitonium scalare has been determined in this study. The complete genome (mtDNA) is 15,140 base pairs (bp) in length and contained of 13 protein-coding genes, two rRNA genes, and 22 tRNA genes. The control region was divided into two parts. The overall base composition of the genome in descending order was 41.1%—T, 28.3%—A, 19.2%—G, and 11.4%—C. In 13 protein-coding genes, COX3, atp6, and nad1 start with ATA. For the stop codon, 11 genes with TAA, COX3, and nad3 end with TAG. Phylogenetic analysis indicated that E. scalare is not closely to any other families and separate into a single branch. This study was the first study that determined the complete mitochondrial genome of Epitonidae species. It would be a supplement for the genetic analysis of E. scalare and promote the phylogenetic of Epitonidae.
Epitonium scalare belongs to the family Gastropoda. It widely distributes in the Indo-Pacific region. It is a common species that can be found in the northwest Pacific Ocean (Richter and Luque Citation2004). The length of adult shells is between 25 mm and 72 mm (Dance Citation2010). It is hermaphroditic species and its spawning period is from June to July. Compared other same genera, the species E. scalare is very attractive with their quite interesting structure, which the whorls of E. scalare are mutual independence and the shell only uses the ribs to hold together (Bouchet et al. Citation2017). Its beautiful shell makes many people would like to use them as the ornamentation (Richter and Luque Citation2004). However, the Epitonidae taxonomy is chaotic and the genetic information of E. scalare is not clear.
Herein, it is the first time that determined the complete mitochondrial genome of E. scalare in this study. The specimen of E. scalare was collected in Zhejiang province, China (122.2°E, 30.3°N) and identified by morphology and deposited in Zhejiang Ocean University. The total DNA extraction was utilized the salting-out method (Aljanabi and Martinez Citation1997) with the muscle. Then, total genomic DNA was diluted to a final concentration of 60–80 ng/μl in 1 × TE buffer and stored at 4 °C in our laboratory at Zhejiang Ocean University for further analysis. The genomic DNA was prepared in 400 bp paired-end libraries. The Illumina HiSeq X Ten platform was used to perform the high-throughput sequence.
The complete mitochondrial genome of E. scalare is 15,140 bp in length (GenBank accession number: MK251987). The complete mitochondrial genome has 13 protein-coding genes, two ribosomal RNA genes, and 22 transfer RNA genes (tRNA). The D-loop region is divided into two parts, one part was between tRNAAsp and atp6 genes; other part was between nad6 and cytb genes. The nucleotide composition for E. scalare is 28.3% of A, 41.1% of T, 11.4% of C, and 19.2% of G. In 13 protein-coding genes, only COX3, atp6 genes and nad1 start with ATA, the others are ATG. For the stop codon, COX3 and nad3 end with TAG, other 11 genes end with TAA. The 16S rRNA is 1308 bp between the tRNA Val and tRNA Leu2, and the 12S rRNA is 858 bp between the tRNAGlu and tRNAVal.
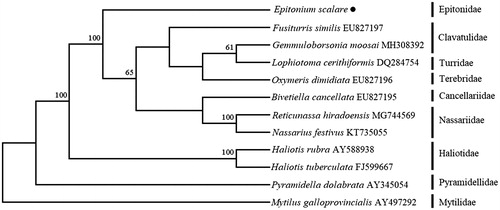
The phylogenetic tree () was constructed based on 13 protein-coding genes of E. scalare and other 11 species using the Neighbor-joining method (Schull Citation1979) in the program Phylip (Felsenstein Citation1989). The tree showed that the E. scalare is not closely to any other families and separate into a single branch. We believe that this result will further supplement the genome information in mitochondria of the family Epitonidae and Facilitate the study on population genetic.
Disclosure statement
The authors declare no competing interests and are the sole authors of the paper. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Aljanabi SM, Martinez I. 1997. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 25:4692–4693.
- Bouchet P, Rocroi J-P, Hausdorf B, Kaim A, Kano Y, Nützel A, Parkhaev P, Schrödl M, Strong EE. 2017. Revised classification, nomenclator and typification of gastropod and monoplacophoran families. Malacologia. 61:1–526.
- Dance SP. 2010. Report on the Linnaean shell collection. Proc Linnean Soc London. 178:1–24.
- Felsenstein J. 1989. PHYLIP-Phylogeny inference package (Version 3.2). Cladistics. 5:164–166.
- Richter A, Luque ÁA. 2004. Epitonium dendrophylliae (Gastropoda: Epitoniidae) feeding on Astroides calycularis (Anthozoa, Scleractinia). J Molluscan Stud. 70:99–101.
- Schull WJ. 1979. Genetic structure of human populations. J Toxicol Environ Health. 5:17–25.