Abstract
Hybridization between birds in wild nature is an interesting phenomenon in the wild, but not common in waterfowl in the wild. In this study, we report the complete mtDNA of the natural hybridization bird between Anser albifrons and Anser fabalis. The complete mtDNA of the hybridization is 16,740 bp in length, contains the typical set of 37 genes, including 13 PCGs, two rRNAs, 22 tRNAs, and a 1177 bp CR. In the 13 PCGs, ATG is generally as the start codon, TAA is the most frequent stop codon, one of three, TAA, TAG, and T-, commonly observed. All tRNAs can be folded into canonical cloverleaf secondary structures except for tRNASer (AGY) and tRNALeu (CUN) missing the “DHU” arm. The new mtDNA sequence contains 12S rRNA and 16S rRNA of rRNAs, separated with tRNAval. The CR is 1177 bp in length, located between tRNAGlu and tRNAPhe. Our phylogenetic trees suggest the hybridization has a close genetic relationship among Anser species.
Hybridization refers to crosses between two different species, genera or higher-ranking through sexual reproduction (Gillespie Citation2004). Hybridization usually occurs between closely related, evolutionarily young taxa with a sympatric or parapatric distribution, interspecific hybrids are bred by mating individuals from two species, normally from within the same genus ). Hybridization between birds in wild nature has been previously reported, avian hybrid especially natural hybridization between birds is an interesting phenomenon in the wild).
The hybridization of Anser albifrons × Anser fabalis has been identified based on morphology and molecular. Trace blood samples from Anser albifrons × Anser fabalis was collected using non-invasive methods at Xuancheng City Forestry Bureau, Anhui province, China in December 2017. The sample was stored at −20 °C at School of Resources and Environmental Engineering, Anhui University (Sample codes is AHU-WB20171202). The DNA sample was stored in the −80 °C in School of Resources and Environmental Engineering, Anhui University. The complete mtDNA sequence of Anser albifrons × Anser fabalis has been assigned with GenBank accession number MG902914.
The complete mtDNA of hybridization is 16,740 bp in length, which is nearly identical to those reported for other Anser birds in length (Liu et al. Citation2013, Citation2014). The overall base composition is: A, 30.20%; C, 32.24%; G, 15.06%; and T, 22.50%. A + T content (52.70%) is higher than C + G content (47.30%), which is similar to other Anser birds (Liu et al. Citation2013, Citation2014). It contains the typical set of 37 genes, including 13 protein-coding genes (PCGs) (ATP6, ATP8, COI-III, ND1-6, ND4L, and Cyt b), two rRNAs (12S rRNA and 16S rRNA), 22 tRNAs, and a putative CR (D-loop). The longest PCGs is ND5 (1818 bp), located between tRNALeu (CUN) and Cyt b. The shortest is ATP8 (168 bp), which is between tRNALys and ATP6. Through the 13 PCGs, ATG is the start codon in eleven of the 13 PCGs, whereas COII and ND5 begin with the nonstandard start codon GTG. The standard stop termination codon TAA occurs in most of the same genes except ND2 and ND6 stop with TAG. Furthermore, AGG terminates the COI gene, AGA terminates the ND5 gene, and the incomplete termination codon T occurs in the ND4 gene. The new mtDNA contains 12S rRNA (988 bp) and 16S rRNA (1612 bp) of rRNAs, located between tRNAPhe and tRNALeu, separated by tRNAVal. All the tRNAs could be folded into the typical cloverleaf structure except for tRNASer (AGY) and tRNALeu (CUN), which lack dihydrouridine (DHU) arms. The CR is located between the tRNAGlu and tRNAPhe genes, which is 1177 bp in the length.
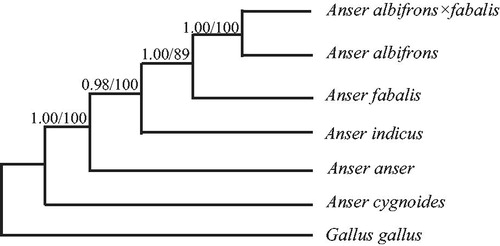
Phylogenetic trees were estimated using ML and BI methods, based on the complete mtDNA of six Anser species, and corresponding Gallus gallus (AY235571) sequence was used as outgroup, sharing similar topologies and high node support values (). Our phylogenetic trees suggest the hybridization has a close genetic relationship among Anser species (Donne-Goussé et al. Citation2002; Gonzalez et al. Citation2009).
Figure 1. Phylogenetic relationships among the six Anser species based on complete mtDNA sequences. Numbers at each node are Bayesian posterior probabilities (left) and maximum likelihood bootstrap proportions (right). The accession number in GenBank of six Anser species in this study: Anser cygnoides (KY767671), Anser anser (NC_011196), Anser indicus (NC_025654), Anser fabalis (HQ890328), Anser albifrons (NC_004539), Anser albifrons × fabalis (MG902914), and Gallus gallus (AY235571).
Nucleotide sequence accession number
The complete mtDNA sequence of Anser albifrons × Anser fabalis has been assigned with GenBank accession number MG902914.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Donne-Goussé C, Laudet V, Hänni CA. 2002. Molecular phylogeny of anseriformes based on mitochondrial DNA analysis. Mol Phylogenet Evol. 23:339–356.
- Gillespie R. 2004. Teaching molecular geometry with the VSEPR model. J Chem Educ. 81: 298–304.
- Gonzalez J, Düttmann H, Wink M. 2009. Phylogenetic relationships based on two mitochondrial genes and hybridization patterns in Anatidae. J Zoo. 279:310–318.
- Liu G, Zhou LZ, Zhang LL, Luo ZJ, Xu WB. 2013. The complete mitochondrial genome of Bean goose (Anser fabalis) and implications for Anseriformes taxonomy. PLoS One. 8:e63334.
- Liu G, Zhou LZ, Li B, Zhang LL. 2014. The complete mitochondrial genome of Aix galericulata and Tadorna ferruginea: bearings on their phylogenetic position in the Anseriformes. PLoS One. 9:e109701.
