Abstract
Aspergillus pseudoglaucus is a xerophilic filamentous fungus which can produce various secondary metabolites. Here, we reported the complete mitochondrial genome sequence of A. pseudoglaucus isolated from Meju, a soybean brick in Korea. Its mitochondrial genome was successfully assembled from raw reads sequenced using MiSeq by Velvet and SOAPGapCloser. Total length of the mitochondrial genome is 53,882 bp, which is third longest among known Aspergillus mitochondrial genomes and encoded 58 genes (30 protein-coding genes including hypothetical ORFs, two rRNAs, and 26 tRNAs). Nucleotide sequence of coding regions takes over 66.6% and overall GC content is 27.8%. Phylogenetic trees present that A. pseudoglaucus is located outside of section Nidulantes. Additional researches will be required for clarifying phylogenetic position of section Aspergillus.
Aspergillus pseudoglaucus (Chen et al. Citation2017), identified in 1929, is a xerophilic filamentous fungus belonging to Aspergillus section Aspergillus. It has been isolated from seafoods and soil (Smetanina et al. Citation2007; Séguin et al. Citation2014). It has also been used as a start culture for Kastuobushi in Japan (Pitt and Hocking Citation2009) and frequently identified from Meju, a soybean brick used for making soy source and soybean paste in Korea (Hong et al. Citation2011). It can produce various metabolites such as benzyl derivatives binding to human opioid or cannabinoid receptors (Gao et al. Citation2011), mycophenolic acid (Mouhamadou et al. Citation2017), and various antibacterial and antifungal compounds (Gao et al. Citation2012). To understand phylogenetic relationship of A. pseudoglaucus, we presented its complete mitochondrial genome.
DNA of A. pseudoglaucus collected from Meju in Korea was extracted using DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Raw data generated using MiSeq, and de novo assembly was conducted using Velvet 1.2.10 (Zerbino and Birney Citation2008). Gap filling was carried out using SOAPGapCloser 1.12 (Zhao et al. Citation2011) after confirming each base using BWA 0.7.17 and SAMtools 1.9 (Li et al. Citation2009; Li Citation2013). Geneious R11 11.0.5 (Biomatters Ltd, Auckland, New Zealand) was used to annotate its mitochondrial genome by comparing with those of Aspergillus luchuensis (MK061298; Park, Kwon, Zhu, Mageswari, Heo, Han, Hong, Citationunder review) and Aspergillus parasiticus (MK124769; Park et al. Citationunder review). Voucher sample was deposited into Korean Agricultural Culture Collection (KACC; Republic of Korea; http://genebank.rda.go.kr/; KACC-93211).
The length of A. pseudoglaucus mitochondrial genome (Genbank accession is MK202802) is 53,882 bp, which is third longest among twelve Aspergillus mitochondrial genomes (Futagami et al. Citation2011; Joardar et al. Citation2012; Xu et al. Citation2018; Park, Kwon, Zhu, Mageswari, Heo, Han, Hong, Citationunder review; Park et al. Citationunder review): The longest is Aspergillus cristatus (77,649 bp; Ge et al. Citation2016) and second longest is Aspergillus egyptiacus (66,526 bp; Xu et al. Citation2018). Aspergillus pseudoglaucus CO1 gene contains eight introns, which is proportion to the length of whole mitochondrial genomes (Joardar et al. Citation2012). In addition, other main protein-coding genes except ND5 have at least one intron, indicating a reason of expansion of A. pseudoglaucus mitochondrial genome (Joardar et al. Citation2012). Moreover, 16 of 20 introns contain partial ORFs, mostly endonuclease. Its mitochondrial genome encoded 58 genes consisting of 30 protein-coding genes including hypothetical ORFs, two rRNAs, and 26 tRNAs. Protein-coding regions takes over 66.6% on genome, and overall GC content is 27.8%, which are similar to those of Aspergillus genus.
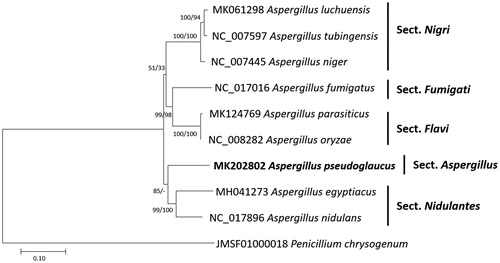
Sequence alignment of nine Aspergillus except A. cristatus, A. egyptiacus, and A. flavus and one Penicillium mitochondrial genome as an outgroup was conducted using MAFFT 7.388 (Katoh and Standley Citation2013). The neighbor joining (10,000 bootstrap repeats) and maximum likelihood (1,000 bootstrap repeats) methods were used for constructing phylogenetic tree using MEGA X (Kumar et al. Citation2018). Phylogenetic trees showed that A. pseudoglaucus is located outside of section Nidulantes and does not form any clades with other species; however, bootstrap supports are not enough (). Additional researches will be required for clarifying phylogenetic position of section Aspergillus.
Figure 1. Neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenetic trees of nine Aspergillus and one Penicillium mitochondrial genome: Aspergillus pseudoglaucus (MK202802, this study), Aspergillus parasiticus (MK124769), Aspergillus luchuensis (MK061298), Aspergillus egyptiacus (MH041273), Aspergillus tubingensis (NC_007597), Aspergillus nidulans (NC_017896), Aspergillus niger (NC_007445), Aspergillus oryzae (NC_008282), Aspergillus fumigatus (NC_017016), and Penicillium chrysogenum (JMSF01000018). The numbers above branches indicate bootstrap support values of neighbor joining and maximum likelihood phylogenetic trees, respectively.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chen AJ, Hubka V, Frisvad JC, Visagie CM, Houbraken J, Meijer M, Varga J, Demirel R, Jurjević Ž, Kubátová A, et al. 2017. Polyphasic taxonomy of Aspergillus section Aspergillus (formerly Eurotium), and its occurrence in indoor environments and food. Stud Mycol. 88:37–135.
- Futagami T, Mori K, Yamashita A, Wada S, Kajiwara Y, Takashita H, Omori T, Takegawa K, Tashiro K, Kuhara S, Goto M. 2011. Genome sequence of the white koji mold Aspergillus kawachii IFO 4308, used for brewing the Japanese distilled spirit shochu. Eukaryotic Cell. 10:1586–1587.
- Gao J, León F, Radwan MM, Dale OR, Husni AS, Manly SP, Lupien S, Wang X, Hill RA, Dugan FM, et al. 2011. Benzyl derivatives with in vitro binding affinity for human opioid and cannabinoid receptors from the fungus Eurotium repens. J Nat Prod. 74:1636–1639.
- Gao J, Radwan MM, León F, Wang X, Jacob MR, Tekwani BL, Khan SI, Lupien S, Hill RA, Dugan FM, et al. 2012. Antimicrobial and antiprotozoal activities of secondary metabolites from the fungus Eurotium repens. Med Chem Res. 21:3080–3086.
- Ge Y, Wang Y, Liu Y, Tan Y, Ren X, Zhang X, Hyde KD, Liu Y, Liu Z. 2016. Comparative genomic and transcriptomic analyses of the Fuzhuan brick tea-fermentation fungus Aspergillus cristatus. BMC Genomics. 17:428.
- Hong S-B, Kim D-H, Lee M, Baek S-Y, Kwon S-W, Samson RA. 2011. Taxonomy of Eurotium species isolated from meju. J. Microbiol. 49:669
- Joardar V, Abrams NF, Hostetler J, Paukstelis PJ, Pakala S, Pakala SB, Zafar N, Abolude OO, Payne G, Andrianopoulos A, et al. 2012. Sequencing of mitochondrial genomes of nine Aspergillus and Penicillium species identifies mobile introns and accessory genes as main sources of genome size variability. BMC Genomics. 13:698.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Preprint arXiv. 13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Mouhamadou B, Sage L, Périgon S, Séguin V, Bouchart V, Legendre P, Caillat M, Yamouni H, Garon D. 2017. Molecular screening of xerophilic Aspergillus strains producing mycophenolic acid. Fungal Biol. 121:103–111.
- Pitt J, Hocking A. 2009. Fungi and food spoilage. 3rd ed. New York: Springer.
- Park J, Kwon W, Zhu B, Mageswari A, Heo I-B, Han K-H, Hong S-B. Under review. Complete mitochondrial genome sequence of the food fermentation fungus, Aspergillus luchuensis. doi:10.1080/23802359.2018.1547160
- Park J, Kwon W, Zhu B, Mageswari A, Heo I-B, Kim J-H, Han K-H, Hong S-B. Under review. Complete mitochondrial genome sequence of an aflatoxin B and G producing fungus, Aspergillus parasiticus. doi:10.1080/23802359.2018.1558126
- Séguin V, Gente S, Heutte N, Vérité P, Kientz-Bouchart V, Sage L, Goux D, Garon D. 2014. First report of mycophenolic acid production by Eurotium repens isolated from agricultural and indoor environments. World Mycotoxin J. 7:321–328.
- Smetanina O, Kalinovskii A, Khudyakova YV, Slinkina N, Pivkin M, Kuznetsova T. 2007. Metabolites from the marine fungus Eurotium repens. Chem Nat Compounds. 43:395–398.
- Xu Z, Wu L, Liu S, Chen Y, Zhao Y, Yang G. 2018. Structure characteristics of Aspergillus egyptiacus mitochondrial genome, an important fungus during the fermentation of dark tea. Mitochondr DNA B. 3:1135–1136.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinform. 12:S2.
