Abstract
In our research, the complete mitochondrial genome of Culicicapa ceylonensise was determined and described, which was the first complete mitogenome reported in the Culicicapa. The complete mitochondrial genome sequence of C. ceylonensise was 16,851 bp in length and contains 13 protein-coding genes (PCGs), two ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, and one control region (D-loop). The overall base composition of the mitochondrial DNA was 29.8% for A, 23.5% for T, 31.8% for C, and 14.9% for G, and the percentage of GC content was 46.7%. The complete mitochondrial genome information of C. ceylonensis will contribute to the phylogenetic studies of the Passeriformes.
Culicicapa ceylonensis was first classified into the subfamily Muscicapidae based on external morphology, reproductive habits, and nesting characteristics. But, the taxonomic status of some branches in the subfamily are still highly controversial (Howard and Moore Citation1991; Voelker and Spellman Citation2004). Cheng (Citation1994) classified the Rhipidura and Culicicapa into the Muscicapidae, but Pasquet et al. (Citation2002) reported that the C. ceylonensis did not belong to the subfamily Muscicapidae. Zheng proposed the Rhipidura to be an independent family of Rhipiduridae (Zheng Citation2005), and Lei et al. (2007) classified the Culicicapa into Rhipiduridae. Molecular studies about C. ceylonensis were limited and the complete mitochondrial genome of C. ceylonensis has not been reported. Here, we first sequenced and assembled the complete mitochondrial genome of C. ceylonensis, the information of the complete mitochondrial genome of C. ceylonensis is useful in the phylogenetic studies of the family Muscicapidae in the future.
Muscle samples of wild C. ceylonensis that died of natural causes were collected from Laojunshan National Nature Reserve, Yibin, Sichuan Province, China (104°00.99’, 28°41.98’). The specimens now were stored in the Museum of Sichuan University. Total genomic DNA was extracted from muscle tissue samples. The complete mitochondrial genome was amplified using PCR with seven pairs of primers designed. The complete mitochondrial genome of C. ceylonensis was a circular DNA molecule with a length of 16,851 bp and has been deposited in GenBank database with an accession number of MH880820. The overall base composition was 29.8% A, 23.5% T, 31.8% C, and 14.9% G, with a GC ratio of 46.7%. The complete mitogenome of C. ceylonensis included 13 protein-coding genes, two ribosomal RNA genes (12S rRNA and 16S rRNA), 22 transfer RNA (tRNA) genes, and one non-coding control region (D-loop). Among these genes, ND6 and eight tRNA genes (tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser, tRNA-Glu, and tRNA-Pro) located on the light strand (L-strand), whereas other genes located on the heavy strand (H-strand).
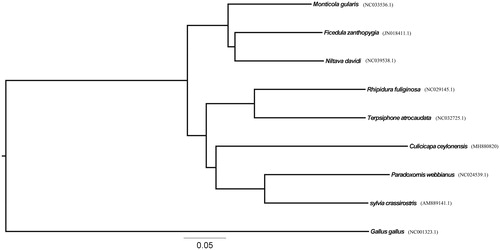
The DNA data of 13 PCGs of C. ceylonensis and other eight Passeriformes species were used for constructing the phylogenetic tree. RAxML (Randomized Axelerated Maximum Likelihood) (Stamatakis Citation2014) is a popular program for phylogenetic analyses of large datasets under maximum likelihood. As indicated by the trees (), the result is similar to the previous study (Cheng Citation1994). The sequence data will be useful for the phylogenetic studies of the family Muscicapidae in the future
Figure 1. A Phylogenetic tree of nine species was constructed based on the data set of 13 concatenated mitochondrial PCGs using RAxML. Sequence data used in the study are the following: Ficedula zanthopygia (JN018411.1), Niltava davidi (NC039538.1), Rhipidura fuliginosa (NC029145.1), Terpsiphone atrocaudata (NC032725.1), sylvia crassirostris (AM889141.1), Monticola gularis(NC033536.1), Paradoxornis webbianus (NC024539.1), Culicicapa ceylonensis (MH880820), and Gallus gallus (NC001323.1).
Acknowledgements
We would like to thank Mr Guo Cai at Sichuan University for assistance in specimens collection in the field.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
Additional information
Funding
References
- Cheng T-H. 1994. A complete checklist of species and subspecies of the Chinese birds. Beijing: Science Press; p. 137–154
- Howard R, Moore A. 1991. A complete checklist of the birds of the world. 2nd ed. London: Academic Press; p. 278.
- Lei X, Lian ZM, Lei FM, Yin ZH, Zhao HF. 2007. Phylogeny of some Muscicapinae genera and species inferred from mitochondrial gene COI sequences. Zool Res. 28:291–296.
- Pasquet E, Cibois A, Baillon F, Erard C. 2002. What are African monarchs (Aves, Passeriformes)? A phylogenetic analysis of mitochondrial genes. C R Biologies. 325:107–118.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Voelker G, Spellman GM. 2004. Nuclear and mitochondrial DNA evidence of polyphyly in the avian superfamily Muscicapoidea. Mol Phylogenet Evol. 30:386–394.
- Zheng GM. 2005. A checklist on the classification and distribution of the birds of China. Beijing: Science Press; p. 235–246.
