Abstract
Manihot esculenta (M. esculenta) is the most important root crop in the world, which has high potential carbohydrate production and adaptability to diverse environments. In this study, we assembled the complete mitochondrial (mt) DNA sequence of M. esculenta into a circular genome of length 682,840 bp, comprising of 32 protein-coding genes, 17 tRNA genes, and three rRNA genes. A neighbour-joining phylogenetic tree was constructed based on 25 plant species and 23 conserved protein-coding genes, suggesting that M. esculenta is evolutionarily close to the Salicaceae plants (Populus tremula and Salix suchowensis) in the same Malpighiales order. The complete mt genome of M. esculenta will provide more desirable information for better understanding the genomic breeding of cassava.
Manihot esculenta, popularly known as cassava or manioc, is a perennial woody shrub in the Euphorbiaceae family, which is cultivated for its starchy storage roots around the tropical and subtropical regions (Wang et al. Citation2014). Because of its high carbohydrate production potential and adaptability to diverse environments, cassava is the most important root crop worldwide and provide major food in South America, Africa, and Asia (Daniell et al. Citation2008). Additionally, cassava is considered as an important reserve of carbohydrates to relieve the global famine because it is highly tolerant to drought and has excellent storage capacity. The complete nuclear and chloroplast genomes of cassava have been assembled; however, the mitochondrial genome has not yet been assembled. The mitochondrion is an endosymbiont-originating organelle, which plays vital roles in cellular ATP production as well as in the regulation of cellular metabolism. Hence, in this study, we primarily report the complete mt genome of M. esculenta and further analyze genomic features.
Sample of M. esculenta used for extraction was generated from a partially inbred line called AM560-2 in Cali, Colombia (Geographic coordinate: 3°30′8″N, 76°21′18″W). The genomic DNA was isolated from fresh young leaves as described previously (Zhang et al. Citation1995) and deposited in CIAT (International Center for Tropical Agriculture). In this study, we assembled the complete mt DNA sequence of M. esculenta into a free-of-gap genome of 682,840 bp according to the reference (Bi et al. Citation2016; Wang et al. Citation2018). The GC content is 44.47%, which is a widespread value in higher plants. After that, we submitted the complete mt genome to GenBank with the accession number of MK176513.
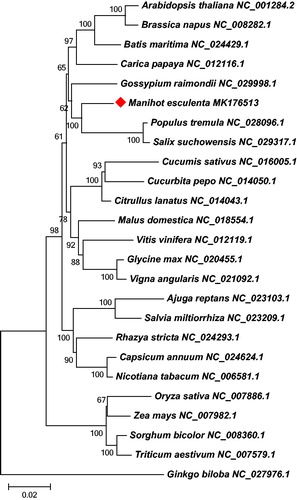
Using the online annotation program MITOFY to annotate the complete mt genome of M. esculenta (Alverson et al. Citation2010), we identified a total of 52 genes in the mt genome, comprising of 32 protein-coding genes, 17 tRNA genes, and 3 rRNA genes. Most of the protein-coding genes use ATG as their start codons, but nad1 and rps4 use ACG as start codons due to a C to U RNA-editing event. Only one gene (mttB) cannot determine its start codon, which is a pervasive case in most higher plants. In the same manner, four different types of stop codons were found in the mt protein-coding genes: TAG (3 genes: atp4, nad4L, and nad7), TGA (11 genes: atp1, atp6, ccmB, ccmC, ccmFn, cob, cox3, nad4, rps1, rps12, and rps13), TAA (17 genes: atp8, atp9, ccmFc, cox2, matR, mttB, nad1, nad2, nad3, nad5, nad6, nad9, rpl5, rpl10, rpl16, rps4, and sdh3) and one specific stop codon of CGA in sdh4 (C to U RNA-editing). The results of stop codons showed that TAA was the most commonly used stop codons, especially for NADH dehydrogenase and Ribosomal proteins (LSU). Phylogenetic analyses were then performed based on an aligned data matrix of 25 plant species and 23 conserved protein-coding genes. A neighbor-joining tree () was then constructed based on the phylogenetic analyses using MEGA6 (Tamura et al. Citation2013) and illustrated that M. esculenta was evolutionarily close to the Salicaceae plants (Populus tremula and Salix suchowensis), and they all belong to the Malpighiales order.
Acknowledgements
We would like to acknowledge the International Center for Tropical Agriculture for their assistance in sampling the material of Manihot esculenta.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Alverson AJ, Wei X, Rice DW, Stern DB, Barry K, Palmer JD. 2010. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol Biol Evol. 27:1436.
- Bi C, Paterson AH, Wang X, Xu Y, Wu D, Qu Y, Jiang A, Ye Q, Ye N. 2016. Analysis of the complete mitochondrial genome sequence of the diploid cotton Gossypium raimondii by comparative genomics approaches. BioMed Res Int. 2016: 18.
- Daniell H, Wurdack KJ, Kanagaraj A, Lee SB, Saski C, Jansen RK. 2008. The complete nucleotide sequence of the cassava (Manihot esculenta) chloroplast genome and the evolution of atpF in Malpighiales: RNA editing and multiple losses of a group II intron. Theor Appl Genet. 116:723–737.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wang W, Feng B, Xiao J, Xia Z, Zhou X, Li P, Zhang W, Wang Y, Møller BL, Zhang P, et al. 2014. Cassava genome from a wild ancestor to cultivated varieties. Nat Commun. 5:5110.
- Wang X, Cheng F, Rohlsen D, Bi C, Wang C, Xu Y, Wei S, Ye Q, Yin T, Ye N. 2018. Organellar genome assembly methods and comparative analysis of horticultural plants. Hortic Res. 5:3.
- Zhang HB, Zhao XP, Ding XL, Paterson AH, Wing RA. 1995. Preparation of megabase-size DNA from plant nuclei. Plant J. 7:175–184.