Abstract
Osmanthus fragrans is valued for its beauty and fragrance. Here, we characterized the complete chloroplast (cp) genome of O. fragrans using next generation sequencing. The circular complete cp genome of O. fragrans is 155,896 bp in length, containing a large single-copy (LSC) region of 86, 521 bp, and a small single-copy (SSC) region of 20943 bp. It comprises 124 genes, including eight rRNA genes, 29 tRNAs genes, and 87 protein-coding genes. The GC content of O. fragrans cp genome is 37.8%. The phylogenetic analysis suggests that O. fragrans is a sister species to Abeliophyllum distichum and Hesperelaea palmeri in Oleaceae.
Keywords:
Osmanthus fragrans Lour. (Oleaceae), also known as the sweet olive, is one of the top 10 traditional flowers in China and has been cultivated for over 2500 years (Xiang and Liu Citation2008). It is a traditional element in landscape gardening and is also a precious raw material for food and spice industries (Yuan et al. Citation2011). But to date, there is still no complete cp genome was characterized for O. fragrans, even for the genera Osmanthus. Here, we characterized the complete cp genome sequence of O. fragrans (GeneBank accession number: MG820121) based on Illumina pair-end sequencing to provide a valuable complete cp genomic resource.
Total genomic DNA was isolated from fresh leaves of O. fragrans grown in Nanjing Forestry University campus in Nanjing, Jiangsu, China. The voucher specimen was deposited at the herbarium of Nanjing Forestry University (accession number NF2010468). Illumina paired-end (PE) DNA library was prepared and sequenced in Nanjing Genepioneer Biotechnologies Inc., Nanjing, China. Raw reads were trimmed using CLC Genomics Workbench v9 and the resultant clean reads were then employed to assemble the cp genome using the program NOVOPlasty (Dierckxsens et al. 2017). The resultant genome was annotated by CpGAVAS (Liu et al. Citation2012). A Neighbor-Joining (NJ) tree with 100 bootstrap replicates was inferred using MEGA version 7 (Tamura et al. Citation2013).
The circular genome of O. fragrans was 155,896 bp in size and contained two inverted repeat (IRa and IRb) regions of 24,216 bp, which were separated by a large single copy (LSC) region of 86,521 bp, and a small single copy (SSC) region of 20943 bp. A total of 124 genes are encoded, including 87 protein-coding genes (80 PCG species), 29 tRNAs gene (24 tRNA species), and eight rRNA genes (four rRNA species). Most of genes occurred in a single copy; however, eight protein-coding genes (ndhB, psbG, rpl2, rpl23, rps7, ycf2, ycf5, and ycf15), five tRNA genes (trnI-CAT, trnL-CAA, trnN-GTT, trnR-ACG, and trnV-GAC), and four rRNA genes (4.5S, 5S, 16S, and 23S) are totally duplicated. A total of seven protein-coding genes (atpF, ndhA, rpoC1, two ndhB, and two rpl2) contained one intron while the other two genes (clpP, ycf3) had two intron each. The overall GC content of O. fragrans genome is 37.8%, and the corresponding values in LSC, SSC and IR regions are 35.8, 33.7, and 43.1%, respectively.
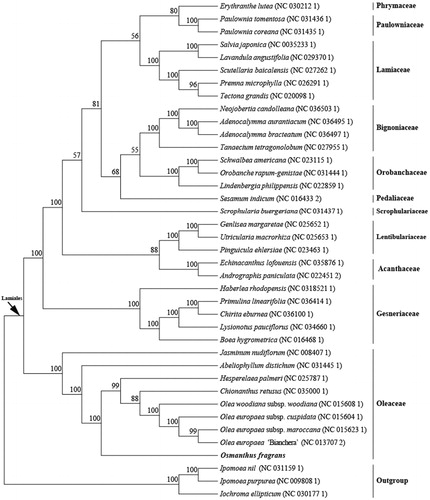
The phylogenetic analysis was conducted based on 36 Lamiales cp genomes and three taxa (Ipomoea nil, Ipomoea purpurea, and Iochroma ellipticum) as outgroups with sequenced cp genomes. We found that O. fragrans was clustered with other families of Lamiales with 100% bootstrap values (). In addition, O. fragrans was highly supported to be a sister species to Abeliophyllum distichum and Hesperelaea palmeri in Oleaceae. Furthermore, this study will pave the way for future research to understand the genomic information of the chloroplasts of the genus Osmanthus and this chloroplast resource could also be utilized on the phylogeny, DNA barcoding, and conservation genetics.
Figure 1. Maximum likelihood tree showing the relationship among Osmanthus fragrans and representative species within the order Lamiales, based on whole chloroplast genome sequences, with three species from Solanales as outgroup. Shown next to the nodes are bootstrap support values based on 1000 replicates.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Xiang QB, Liu YL. 2008. An illustrated monograph of the sweet Osmanthus cultivars in China. Hangzhou: Zhejiang Science and Technology Press.
- Yuan WJ, Han YJ, Dong MF, Shang FD. 2011. Assessment of genetic diversity and relationships among Osmanthus fragrans cultivars using AFLP markers. Electron J Biotechn. 14:e1–e9.
