Abstract
The mitochondrial genome of a blacktip sardinella, Sardinella melanura was determined by high-throughput sequencing (HTS) technique. The mitogenome of S. melanura (16,646 bp), encoded the canonical 37 mitochondrial structural genes (13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes). The gene arrangement of S. melanura was identical to its relative species including Sardinella lemuru, Sardinella longiceps and Sardinella gibbosa. Except for COX1 (GTG), all the other PCGs showed the typical ATG as start codon. Incomplete stop codons were identified in ND2, COX2, COX3, ND3, ND4, and Cytb. The phylogenetic tree result showed that S. melanura was most closely related to S. jussieu, S. lemuru, and Sardinella maderansis with 83% identity, respectively.
The region of Indo-west Pacific ocean is known for the high diversity of clupeid species, some of which are still taxonomically ambiguous awaiting the clear description (Thomas et al. Citation2014). The blacktip sardinella, Sardinella melanura is a marine amphidromous clupeid species, which is commercially important in South-west and South-east Asia (Whitehead et al. Citation1985; Pedrosa-Gerasmio et al. Citation2015; Nelson et al. Citation2016). For the effective and scientific management of S. melanura with its relative species sharing the habitats, its genetic information should be required. In this study, the complete mitogenome of S. melanura was determined by Illumina MiSeq platform and its phlylogenetic relationship with other clupeid species was analyzed.
The specimen was collected from the coastal water in Banyuwangi, East Java, Indonesia (8°12′07.52″S 114°23′07.18″E) and stored at University of Airlangga, Indonesia. Species identification of the specimen was confirmed by both its keys morphological characteristics and 100% nucleotide sequence identity of the COI region to the database (KX223945). The mitochondrial DNA extracted by a commercial kit (Abcam, Cambridge, UK ) was further fragmented into the smaller sizes (∼350 bp) by Covaris M220 Focused-Ultrasonicator (Covaris Inc., Woburn, MA, USA). A library for sequencing was constructed by TruSeq® RNA library preparation kit V2 (Illumina Inc., San Diego, CA, USA) and its quality and the quantity was analyzed by 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). DNA sequencing was performed by Illumina MiSeq sequencer (2 × 300 bp pair ends). The complete mitogenome was assembled by Geneious v 11.0.2 (Kearse et al. Citation2012) and tRNA structures were predicted by ARWEN (Laslett and Canbäck Citation2008).
The complete mitochondrial genome of S. melanura (MH995532) was 16,646 bp in length, which consisted of 13 protein-coding genes (PCGs), 22 tRNAs, 2 ribosomal RNAs (12S and 16S), and 2 non-coding regions including the origin of light strand replication (OL) and the putative control region (D-Loop). The gene arrangement of S. melanura was identical to its relatives, Sardinella lemuru (Jiang et al. Citation2018), Sardinella longiceps, and Sardinella gibbosa (Sebastian et al. Citation2017). Twelve PCGs were on the heavy strand, except for the ND6. Three conserved tRNA clusters (IQM, WANCY, and HSL) were also well conserved (Satoh et al. Citation2016). All the tRNAs were predicted to form the typical stem-loop secondary structures as shown in its relative species S. lemuru. Two non-coding regions, OL and D-Loop was located between tRNAAsn and tRNACys and between tRNAPro and tRNAPhe, respectively. Except for COX1 (GTG), all the other PCGs showed the typical ATG as start codon. Incomplete stop codons were identified in ND2, COX2, COX3, ND3, ND4, and Cytb.
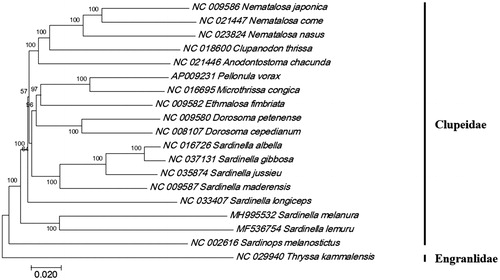
The phylogenetic tree of S. melanura with 17 clupeid species was constructed by MEGA7 with minimum evolutionary (ME) algorithm (Kumar et al. Citation2016). This result showed that S. melanura was most closely related to S. jussieu (Sektiana et al. Citation2017), S. lemuru, and Sardinella maderansis with 83% identity ().
Figure 1. Phylogenetic tree of Sardinella melanura within family Clupeidae. Phylogenetic tree of the mitochondrail complete genomes in Clupeidae was constructed by MEGA7 software by Minimum Evolution (ME) algorithm with 1000 bootstrap replications. The number at each node is bootstrap probability. GenBank Accession numbers were shown followed each scientific name.
Disclosure statement
The authors report that they have no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Jiang H, Gong L, Liu L, Liu B, Lü Z. 2018. The complete mitochondrial genome of Sardinella lemuru (Clupeinae, Clupeidae, Clupeoidei) and phylogenetic studies of Clupeoidei. Mitochondrial DNA B. 3:50–52.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Nelson JS, Grande TC, Wilson MV. 2016. Fishes of the world. Hoboken, New Jersey, USA: John Wiley & Sons.
- Pedrosa-Gerasmio IR, Agmata AB, Santos MD. 2015. Genetic diversity, population genetic structure, and demographic history of Auxis thazard (Perciformes), Selar crumenophthalmus (Perciformes), Rastrelliger kanagurta (Perciformes) and Sardinella lemuru (Clupeiformes) in Sulu-Celebes Sea inferred by mitochondrial DNA sequences. Fish Res. 162:64–74.
- Satoh TP, Miya M, Mabuchi K, Nishida M. 2016. Structure and variation of the mitochondrial genome of fishes. BMC Genomics. 17:719–738.
- Sebastian W, Sukumaran S, Zacharia P, Gopalakrishnan A. 2017. The complete mitochondrial genome and phylogeny of Indian oil sardine, Sardinella longiceps and Goldstripe Sardinella, Sardinella gibbosa from the Indian Ocean. Conservation Genetics Resources. 10:1–5.
- Sektiana SP, Andriyono S, Kim H-W. 2017. Characterization of the complete mitochondrial genome of Mauritian sardinella, Sardinella jussieu (Lacepède, 1803), collected in the Banten Bay, Indonesia. Fish Aquatic Sci. 20:26.
- Thomas RC Jr, Willette DA, Carpenter KE, Santos MD. 2014. Hidden diversity in sardines: genetic and morphological evidence for cryptic species in the goldstripe sardinella, Sardinella gibbosa (Bleeker, 1849). PLoS One. 9:e84719.
- Whitehead P, Nelson G, Wongratana T. 1985. FAO species catalogue. Vol. 7-Clupeiod fishes of the world. An annotated and illustrated catalogue of the herrings. Sardines, Pilchards, Sprats, Shad, Anchovies and Wolf Herring. Part 1-Chirocentridae Clupleidae and Pristigasteridae. FAO Fisheries Synopsis. 125:90–113.
