Abstract
Quercus mongolica is a major tree species in East Asian hardwood forests. In this study, we assembled a complete chloroplast (cp) genome of Q. mongolica based on next-generation sequencing data. The cp genome was 161,295 bp, with a large single-copy region (90,583 bp), a small single-copy region (19,060 bp), and two short inverted repeat regions (25,826bp). The genome contains 121 genes, including 85 protein-coding genes, eight rRNA genes, and 29 tRNA genes. Phylogenetic analyses based on cp genome sequences revealed that Q. mongolica was grouped with species from section Quercus.
The Mongolian oak (Quercus mongolica) is a dominant tree species in East Asian hardwood forests (Wang, et al. Citation1985). It plays a key role in windbreak, water conservation, fire-prevention, and in the economic and ecosystemic development in China, Russia, Korea, and Japan (Yang et al. Citation2016). However, only a few genomic resources have been explored in this species (Zeng et al. Citation2010; Zeng et al. Citation2015; Yang et al. Citation2016). Here, we sequenced and analyzed the chloroplast (cp) genome of Q. mongolica based on the next-generation sequencing data. The objectives of this study were to characterize the complete cp genome sequence of Q. mongolica as a resource for future genetic studies on this and other related species.
Fresh leaves of Q. mongolica were collected from a maturate tree in the Institute of Botany, CAS (Beijing, China). After DNA extraction, a library with the insertion size of 350 bp was constructed, and high-throughput DNA sequencing (pair-end 150bp) was performed on an Illumina X10 platform. In total, 8G raw reads were obtained for de novo assembly of cp genome in NOVOPlasty (Dierckxsens et al. Citation2017). A ribulose-bisphosphate carboxylase (RBCL) gene sequence from Quercus rubor was used as seed sequence, and the whole cp genome sequence of Q. rubor was used as a reference to resolve the inverted repeat in the cp genome of Q. mongolica. Default settings were used for other parameters. The assembled cp genome was annotated using CpGAVAS (http://124.17.107.12/0506/cpgavas/analyzer/home).
The whole cp genome of Q. mongolica was 161,295 bp in length, with two short inverted repeat (IRa and IRb) regions of 25,826 bp, a large single-copy (LSC) region of 90,583 bp, and a small single-copy (SSC) region of 19,060 bp. The overall GC content of cp genome is 36.8%. The cp genome of Q. mongolica contained 122 genes, including 85 protein-coding genes, eight rRNA genes, and 29 tRNA genes. Most of these genes did not contain intron, 10 genes contained one intron, and two genes (clpP and ycf3) contained two introns. All genes occurred as a single copy, except that 21 genes were duplicated in IR regions.
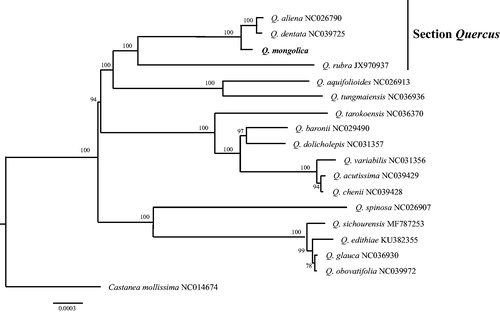
A phylogenetic analysis was carried out using cp genome sequences of Q. mongolica and other 16 species within genus Quercus (). The sequences were aligned using ClustalW v.2.1 (Larkin et al. Citation2007), and a neighbour-joining tree was constructed using MEGA X (Kumar et al. Citation2018) with Castanea mollissima as an outgroup. Phylogenetic analysis revealed that Q. mongolica was grouped with species of section Quercus (bootstrap = 100), in consistent with the previous study in Oaks (Pearse and Hipp Citation2009). In conclusion, this complete cp genome would establish a solid foundation for future genetic studies in Q. mongolica and its relative species.
Data availability
The chloroplast genome of the Q. mongolica was submitted to Genbank under accession number: MK564083.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and clustal X version 2.0. Bioinformatics. 23:2947–2948.
- Pearse IS, Hipp AL. 2009. Phylogenetic and trait similarity to a native species predict herbivory on non-native oaks. Proc Natl Acad Sci USA. 106:18097–18102.
- Wang LM, Ren XW, Liu YQ. 1985. Geographic distribution of deciduous oaks in China. J Beijing Fores Uni. 2:57–69.
- Yang J, Di XY, Meng X, Feng L, Liu Z, Zhao GF. 2016. Phylogeography and evolution of two closely related oak species (Quercus) from north and northeast China. Tree Genet Genomes. 12:89.
- Zeng YF, Liao WJ, Petit RJ, Zhang DY. 2010. Exploring species limits in two closely related Chinese oaks. PloS One. 5:e15529
- Zeng YF, Wang WT, Liao WJ, Wang HF, Zhang DY. 2015. Multiple glacial refugia for cool-temperate deciduous trees in northern East Asia: the Mongolian oak as a case study. Mol Ecol. 24:5676–5691.