Abstract
Orthriophis moellendorffi (Colubridae, Serpentes) mainly inhabits in an altitude of 50–300 meters in south China and also in Vietnam. The complete mitogenome sequence of O. moellendorffi is 17,191 bp in length, including 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and a D-loop region. The base composition of mitogenome was 35.4%A, 26.2% C, 12.3% G, and 26.1% T. The ND6 subunit gene and eight tRNA genes are encoded on the L-strand, the others are encoded on the H-strand.
Orthriophis moellendorffi (Colubridae, Serpentes), formerly known as the Elaphe moellendorffi is distributed in Vietnam, Guangdong, and Guangxi Province and other places in China, which mainly inhabits at an altitude of 50–300 meters, living in the foot of rock/rocky grass, as well as roadside, ditch and small river beside gravel grass. The total length of O. moellendorffi is up to 200 cm, the head is similar to the pear-like shape with a reddish-brown color, the lips and the body are gray.
Yet, there were a few studies on the phylogenetic position of O. moellendorffi. In the present study, the complete mtDNA of O. moellendorffi was amplified and sequenced using 23 primer pairs and then submitted to GenBank (accession number MK408450). This study will provide preliminary molecular data for the phylogenetics and evolution study of this species.
In the study, the specimen was stored in the conservation biology laboratory of Anhui Normal University in Wuhu, Anhui, China. The muscle tissues were used to extract the whole genomic DNA using a standard proteinase-k/phenol-chloroform protocol (Sambrook and Russell Citation2001). The entire mitogenome was amplified using 23 pairs of primers by polymerase chain reaction. For probing the phylogenetic status of O. moellendorffi, nine mitochondrial genomes of Colubridae species and an outgroup (Deinagkistrodon acutus) were collected from GenBank. Phylogenetic tree was constructed by the maximum likelihood (ML) method implemented in the Mega 5 software (Tamura et al. Citation2011). The best-fitting nucleotide substitution model (GTR + I+G) was selected via JModelTest 0.1.1 (Campos et al. Citation2013). Support for the internal branches was evaluated with 1000 bootstrap replicates. Other parameters were used as default.
The complete mitogenome sequence of O. moellendorffi is 17,191 bp in length, which includes 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and a D-loop region. The base composition of the mitogenome was 35.4%A, 26.2%C, 12.3%G, and 26.1%T. The ND6 subunit gene and eight tRNA genes were encoded on the L-strand, the others were encoded on the H-strand. In 13 protein-coding genes, the shortest one was ATP8 gene (159 bp) and the longest one was the ND5 gene (1767 bp). For most protein-coding genes, they take ATG as start codons, while COX I regarded GTG as start codons. The D-loop of O. moellendorffi mtDNA was 1026 bp in length.
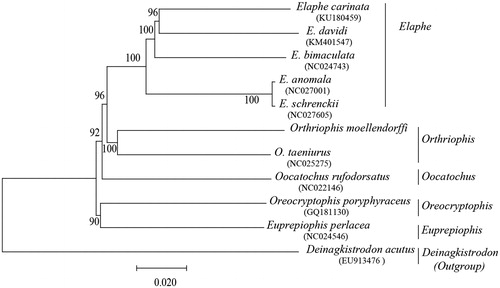
The phylogenetic tree divided into five well-supported clades (), which corresponds to five genera (Elaphe, Orthriophis, Oocatochus, Oreocryptophis, and Euprepiophis). The topology of the phylogenetic tree was largely consensus with previous studies (Pyron et al. Citation2013; Chen et al. Citation2014), the result pointed out that the relationship of O. moellendorffi and O. taeniurus was very close to each other, which are species of Orthriophis, forming a paraphyletic group with Elaphe, Oocatochus, Oreocryptophis, and Euprepiophis. Our research supports the current Colubridae classification system, and the mitochondrial genome of this species will provide basic theoretical data for the systematic study of the Colubridae.
Figure 1. The Mega tree and maximum-likelihood (ML) tree among 10 species based on the whole mitogenome. Numbers at the nodes are bootstrap values of the Mega and ML analysis. The GenBank accession number of species are Elaphe carinata (KU180459), E. davidi (KM401547), E. bimaculata (NC024743), E. anomala (NC027001), E. schrenckii (NC027605), Orthriophis taeniurus (NC025275), Oocatochus rufodorsatus (NC022146), Oreocryptophis porphyraceus (GQ181130), Euprepiophis perlacea (NC024546), Deinagkistrodon acutus (EU913476).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Campos R, Botto-Mahan C, Coronado X, Catala SS, Solari A. 2013. Phylogenetic relationships of the Spinolai complex and other Triatomini based on mitochondrial DNA sequences (Hemiptera: Reduviidae). Vector Borne Zoonotic Dis. 13:73–76.
- Chen X, Jiang K, Guo P, Huang S, Rao D, Ding L, Takeuchi H, Che J, Zhang YP, Myers EA, Burbrink FT. 2014. Assessing species boundaries and the phylogenetic position of the rare Szechwan ratsnake, Euprepiophis perlaceus (Serpentes: Colubridae), using coalescent-based methods. Mol Phylogenetics Evol. 70:130–136.
- Pyron RA, Burbrink FT, Wiens JJ. 2013. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol Biol. 13:93.
- Sambrook J, Russell DW. 2001. Rapid isolation of yeast DNA.In: Sambrook J & Russell DW, eds. Molecular Cloning, aLaboratory Manual, New York: Cold Spring Harbor Lab-oratory Press, pp. 631–632.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.
