Abstract
Solanum chacoense is a wild tuber-bearing species and an important resource used as inbred line to improve common scab resistance in interspecies transfer to cultivated potato (S. tuberosum). In this study, we report the complete chloroplast genome sequence of S. chacoense using Illumina sequencing technology. The complete chloroplast genome was 155,528 bp in length and comprised a pair of inverted repeats (IR) regions of 25,592 bp separated by a large single-copy (LSC) region of 86,178 bp and a small single-copy (SSC) region of 18,586 bp. The GC content was 37.9% overall. The genome contained 132 genes including 87 protein-coding genes (PCGs), 37 tRNAs and 8 rRNAs. Phylogenetic analysis based on eight complete chloroplast genomes showed a sister-group relationship between S. chacoense and S. commersonii.
Wild potatoes are broadly distributed in the America from the southwestern United States to the Southern Cone of South America (Spooner et al. Citation2014). They possess rich genetic variations which are readily available to potato breeder (Hawkes Citation1958; Hanneman Citation1989; Jansky et al. Citation2014). As a member of wild relative potato, Solanum chacoense, is identified to be high resistance to common scab, Colorado Potato Beetle, verticillium wilt, and an important source of potato breeding (Lynch et al. Citation1997; Ronning et al. Citation2000; Jansky et al. Citation2018b). A clone of S. chacoense, 524-8, was identified as highly resistant to common scab and used to create common scab resistant tetraploid potato clone (Jansky et al. Citation2018a). In addition, it was used to create self-compatible potato clone M6 which is a vigorous and fertile inbred line. M6 is a useful tool in diploid breeding, thus the whole genome of this clone was sequenced for effective utilization (Jansky et al. Citation2014; Leisner et al. Citation2018). In this study, we generated a chloroplast genome sequence, assembled, and characterized the complete nucleotide sequence of S. chacoense in chloroplast genome.
The S. chacoense (S-695) was provided by Huazhong Agricultural University, China. A pair-end library was constructed with total genomic DNA using NovaSeq platform (Illumina, San Diego, CA). Low-quality reads were filtered and about 4.0 Gb of clean reads were assembled using GetOrganelle pipeline (Jin et al. Citation2018). In this pipeline, chloroplast genome of S. stoloniferum was set as a reference sequence. Plastid-like reads were treated as ‘baits’ and Bowtie 2 (Langmead and Salzberg Citation2012) was used to map the baits sequence to the reference sequence. It got more plastid-like reads with multiple extension iterations until no more chloroplast reads can be extracted. Then, the total plastid-like reads were de novo assembled into a FASTA Graph (‘fastg’) using SPAdes (Bankevich et al. Citation2012). The complete circular chloroplast genome was finalized through a visual software, Bandage (Wick et al. Citation2015). The annotation of chloroplast genome was performed using a Perl script, PGA (https://github.com/quxiaojian/PGA). To determine the position of S. chacoense in section Petota, seven plastomes of Solanum was downloaded from GenBank including five wild potato relatives and two outgroups, S. lycopersicum and S. nigrum. Whole chloroplast genome sequences were aligned in MAFFT v. 7 (Katoh and Standley Citation2013). The phylogenetic relationship of eight solanum species was reconstructed using RAxML v.8.1.1179 (Stamatakis Citation2014) at the XSEDE Teragrid of CIPRES science Gateway and adopting 1000 replicates of rapid bootstrap with GTRGAMMA substitution model (Miller et al. Citation2010).
Figure 1. Maximum likelihood phylogenetic tree of S. chacoense based on eight complete chloroplast genome sequences using S. lycopersicum and S. nigrum as outgroups. Bootstrap values are given above branches based 1000 replicates.
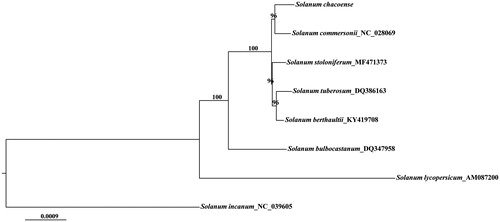
The complete chloroplast genome of S. chacoense (GenBank accession no. MK398247) was 155,528 bp in length consisting of a pair of inverted repeats (IR) regions of 25,592 bp separated by a large single-copy (LSC) region of 86,178 bp and a small single-copy (SSC) region of 18,586 bp. The GC content was 37.9% overall. The complete chloroplast genome comprised 132 genes including 87 protein-coding genes (PCGs), 37 tRNAs, and 8 rRNAs. Furthermore, multiple copies of seven PCGs, four rRNAs and seven tRNAs genes were completely duplicated within IR. Within maximum likelihood phylogenetic tree, S. chacoense and S. commersonii formed a sister-group clade ().
Acknowledgments
We appreciated the help of Li Zhong and Jian-Jun Jin on the bioinformatic analyses.
Disclosure statement
The author reports no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Hanneman RE. 1989. The potato germplasm resource. Amer Potato J. 66:655–667.
- Hawkes JG. 1958. Significance of wild species and primitive forms for potato breeding. Euphytica. 7:257–270.
- Jansky SH, Douches DS, Haynes K. 2018a. Germplasm release: three tetraploid potato clones with resistance to common scab. Am J Potato Res. 95:178–182.
- Jansky SH, Douches DS, Haynes K. 2018b. Transmission of scab resistance to tetraploid potato via unilateral sexual polyploidization. Am J Potato Res. 95:272–277.
- Jansky SH, Chung YS, Kittipadukal P. 2014. M6: a diploid potato inbred line for use in breeding and genetics research. J Plant Regist. 8:195–199.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. 4:256479.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357.
- Leisner CP, Hamilton JP, Crisovan E, Manrique-Carpintero NC, Marand AP, Newton L, Pham GM, Jiang J, Douches DS, Jansky SH, et al. 2018. Genome sequence of M6, a diploid inbred clone of the high-glycoalkaloid-producing tuber-bearing potato species Solanum chacoense, reveals residual heterozygosity. Plant J. 94:562–570.
- Lynch DR, Kawchuk LM, Hachey J, Bains PS, Howard RJ. 1997. Identification of a gene conferring high levels of resistance to Verticillium Wilt in Solanum chacoense. Plant Dis. 81:1011–1014.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE), 14 Nov. 2010, New Orleans, LA pp 1–8.
- Ronning CM, Kowalski SP, Sanford LL, Stommel JR. 2000. Geographical variation of solanidane aglycone glycoalkaloids in the wild potato species Solanum chacoense Bitter. Genet Resour Crop Ev. 47:359–369.
- Spooner DM, Ghislain M, Simon R, Jansky SH, Gavrilenko T. 2014. Systematics, diversity, genetics, and evolution of wild and cultivated potatoes. Bot Rev. 80:283–383.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31:3350–3352.
