Abstract
The Chinese kukri snake Oligodon chinensis belongs to family Colubridae and is distributed in southern China and North Vietnam. In this study, the total mitochondrial genome of O. chinensis was determined using next-generation sequencing.
It is a circular molecule of 17,146 bp in length and contains 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and 2 control regions (CR1 and CR2), with a base composition of A 33.6%, G 12.2%, T 25.6%, and C 28.6%. Interestingly, a special 136 bp non-coding region, which was identified as the repeated sequence in CR1 and CR2. Oligodon chinensis lay a basal position within Colubrinae (Clade A). Ptyas mucosa was the nearest sister to Elaphe carinata and they were clustered in other species of Elaphe. The molecular data presented here would be useful for further study of O. chinensis.
The Chinese Kukri Snake Oligodon chinensis belongs to the family Colubridae and the species is found in southern China and North Vietnam (Orlov et al. Citation2000; IUCN Citation2012). This nocturnal, terrestrial snake is a generalist that inhabits plains, hills, forests, agricultural fields, and rural villages with an elevation ranging from 100 m to 1500 m.
Until now, only partial fragments of mitochondrial DNA (12S, Cox1, ND4, Cytb) and a nuclear gene (c-mos) were determined. Here, we determined the complete mitochondrial genome of O. chinensis, with the individual (JAF01), captured in Fuheyuan Provincial Natural Reserve (latitude: 26.634°N, longitude: 116.380°E), Guangchang county, Jiangxi province, to provide reference information for further study on this species.
Total genomic DNA was extracted from liver tissue of JAF01 with the DNeasy tissue kit (Qiagen, Hilden, Germany) following the manufacturer's protocol. The mitochondrial genome sequence was obtained by next-generation sequencing. The library was sequenced by General GeneTest Company (Beijing, China) using Ion Torrent platform. The de novo assembly of the mitochondrial contigs was conducted by Assembler SPAdes 3.13.0 (Bankevich et al. Citation2012).
The complete mitochondrial genome of O. chinensis (GenBank Accession Number MK347418, 17,146 bp) contains 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and 2 control regions (CR1 and CR2). Interestingly, the identical non-coding sequence (136 bp) is repeated in CR1 and CR2. The whole base composition of the mitochondrial genome is showed as follows: A 33.6%, G 12.2%, T 25.6%, C 28.6%; with an A + T-rich pattern of the vertebrate mitochondrial genomes (Jang and Hwang Citation2011; Xu et al. Citation2015; Zhou et al. Citation2016).
Total length of the 13 protein-coding genes is 11,265 bp, all of which are encoded on the heavy strand except for Nd6 in the light strand. All of the protein-coding genes initiate with the orthodox ATG start codon except for Nd1, Nd2, Cox1, Nd3, Nd6, which begin with ATC, ATT, GTG, ATC, and ATA, respectively. Four types of stop codons are used by the coding genes, including TAA for Atp8, Atp6, Nd4l, Nd4, AGG for Nd5, Nd6, AGA for Cox1 and an incomplete stop codon T for Nd1, Nd2, Cox2, Cox3, Nd3, and Cytb.
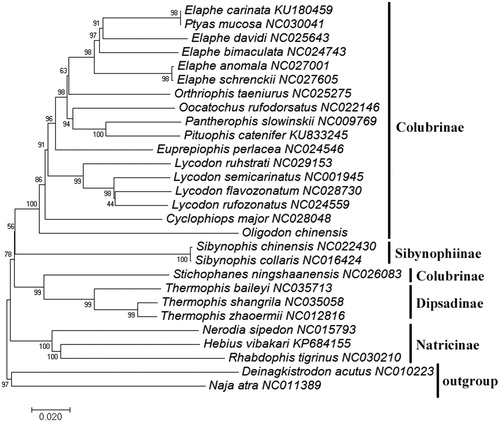
A phylogenetic tree () of mitochondrial genomes analyses of 26 speices snakes of Colubridae and plus two outgroup species, Deinagkistrodon acutus (NC010223) and Naja atra (NC011389), was constructed based on the NJ method. The results indicated that most of the subfamilies were monophyly to the exclusion of Colubrinae (). Oligodon chinensis lay a basal position within Colubrinae (Clade A). Ptyas mucosa was the nearest sister to Elaphe carinata and they were clustered in other species of Elaphe (), which was consistent with the previous study (Zhou et al. Citation2016).
So far, the species status of O. chinensis within Colubridae is unclear. The complete mitochondrial genome of O. chinensis we determined would be useful in systematics and population genetics.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Jang KH, Hwang UW. 2011. Complete mitochondrial genome of the black-headed snake Sibynophis collaris (Squamata, Serpentes, Colubridae). Mitochondrial DNA. 22:77–79.
- IUCN. 2012. IUCN Red List of Threatened Species (ver. 2012.1). Available at:http://www.iucnredlist.org. (Accessed: 19 June 2012).
- Orlov NL, Murphy RW, Papenfuss TJ. 2000. List of snakes of Tam-Dao mountain ridge (Tonkin, Vietnam). Russian Journal of Herpetology. 7: 69–80.
- Xu CZ, Mu YS, Kong QR. 2015. Sequencing and analysis of the complete mitochondrial genome of Elaphe davidi (Squamata: Colubridae). Mitochondrial DNA. 27:1–2.
- Zhou B, Ding CH, Duan YB, Hui G. 2016. The complete mitochondrial genome sequence of Ptyas mucosus. Mitochondrial DNA B. 1:193–194.