Abstract
Pomatosace filicula (Primulaceae) is an alpine biennial endemic to the Qinghai-Tibet Plateau, which is regarded as a kind of traditional Tibetan medicine. We assembled the whole chloroplast genome sequence of this endangered species based on Illumina pair-end sequencing data. The complete chloroplast genome was estimated to be 151,277 bp in length, comprising a large single copy region of 82,039 bp and a small single copy region of 23,848 bp, separated by a pair of 22,695 bp inverted repeat regions. The genome contained 121 genes, including 83 protein-coding genes (74 PCG species), 29 tRNA genes (24 tRNA species), and eight ribosomal RNA genes (four rRNA species). The overall AT content of P. filicula chloroplast genome is estimated to be 62.59%. This genetic resource will facilitate further conservation genetic studies of P. filicula based on designing new chloroplast DNA markers.
Pomatosace filicula (Primulaceae) is an alpine herb endemic to the Qinghai-Tibet Plateau (QTP), which distributed in the disturbed grounds in the alpine meadow ecosystems between 3800 and 4800 m a.s.l. in the northeastern QTP (Wang et al. Citation2004). This species is regarded as a Tibetan folk medicine for treatment of rheumatic arthritis, fever, and haemorrhage. However, it becomes endangered as a threatened species in China because of being over-collected by the local Tibetans.
A well-known chloroplast genome will facilitate further designing more cpDNA markers to monitor changes of the population size of this species, which will contributed to the scientific formulation of protection strategy.
We collected fresh leaves from a single individual of this species from Yushu (N33°34′, E96°02′) and dried them immediately by silica gels. Voucher specimens were deposited in the Sichuan University Herbarium. We extracted the total genomic DNA with the modified CTAB method following Yang et al. (Citation2016). We then constructed 300 bp library and finished whole-genome sequencing with 150 bp pair-end reads using the Illumina Hiseq Platform (Illumina, San Diego, CA). We obtained about 10 million high-quality clean reads. We then assembled the chloroplast genome assembly using NOVOPlasty software (Dierckxsens et al. Citation2016). We linked the resulting contigs based on the overlapping regions after being aligned to Primula veris (Zhou Citation2016). We visualized the genome by Geneious version 8.0.5 (Kearse et al. Citation2012). We performed gene annotation firstly with DOGMA (Wyman et al. Citation2004) and CpGAVAS (Liu et al. Citation2012), then corrected manually with the Geneious (Kearse et al. Citation2012). We generated a physical map of the chloroplast genome by CpGAVAS (Liu et al. Citation2012). The complete chloroplast genome sequence together with gene annotations were submitted to GenBank under the accession number KY612322.
The chloroplast genome sequence of P. filicula is 151,277 bp in length. It comprises two inverted repeat regions of 22,695 each separated by a large single-copy and a small single copy region of 82,039 bp and 23,848 bp, respectively (). We annotated a total of 121 genes, comprising 83 protein-coding genes (74 PCG species), 29 tRNA genes (24 tRNA species), and eight ribosomal RNA genes (four rRNA species). The overall AT content of P. filicula chloroplast genome is estimated to be 62.59%. Compared with the cp genome of Primula veris (Zhou Citation2016) from the same family Primulaceae, most genes are conserved and intergeneric regions show more variations.
Figure 1. Phylogenetic relationship of the P. filicula chloroplast genome with nine previously reported complete chloroplast genomes.
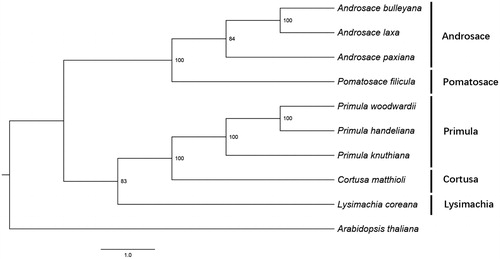
We studied the phylogenetic relationship among P. filicula and nine other previously reported complete chloroplast genomes including eight species from Primulaceae and Arabidopsis thaliana as an outgroup. Alignment was conducted using MAFFT (Katoh and Standley Citation2013). The maximum likelihood phylogenetic tree was built using RAxML (Stamatakis Citation2014) with bootstrap set to 1000.
The detailed population genetic information would be fundamental to formulate potential new conservation and management strategies for this important endangered medicinal species in the Qinghai-Tibet Plateau ().
Table 1. Accession numbers of chloroplast genomes in phylogenetic tree.
Acknowledgments
All calculations were carried out at National Supercomputing Center in Shenzhen.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Liu C, Shi LC, Zhu YJ, Chen HM, Zhang JH, Lin XH, Guan XJ. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genom. 13:715.
- Sato S, Nakamura Y, Kaneko T, et al. 1999. Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Research. 6:283–290.
- Son O, Park SJ. 2014. Complete chloroplast genome sequence of Lysimachia coreana (Primulaceae). Mitochondrial DNA. 27:1–3.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Ting R, Yanci Y, Tao Z, et al. 2018. Comparative Plastid Genomes of Primula Species: Sequence Divergence and Phylogenetic Relationships. International Journal of Molecular Sciences. 19:1050.
- Wang YJ, Li XJ, Hao G, Liu JQ. 2004. Molecular phylogeny and biogeography of Androsace (Primulanceae) and the convergent evolution of cushion morphology. Acta Phytotaxonomica Sinica. 42:481–499.
- Wang J, Zhang R, Ren T, et al. 2017. Characterization of the complete chloroplast genome of the Cortusa matthioli subsp. pekinensis (Primulaceae). Conservation Genetics Resources. 9:603–605.
- Wyman SK, Jansen RK, Boore J. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yang WL, Wang WW, Zhang L, Chen ZY, Guo XY, Ma T. 2016. Characterization of the complete chloroplast genome of Idesia polycarpa. Conservation Genet Resour. 8:271–273.
- Zhou T. 2016. Characterization of the complete chloroplast genome sequence of Primula veris (Ericales: Primulaceae). Conversation. Genet Resour. 8:455–458.
