Abstract
The complete mitochondrial genomic DNA of Cyprinus carpio var. Guilin was 16,582 bp in length, including 22 transfer RNA genes, 13 protein-coding genes, 2 ribosomal RNA genes, and a D-loop region. Phylogenetic analysis indicated that it was most closely related to Cyprinus carpio isolate Oujiang. The results will provide a reference for further studies on the genetic relationships and evolution process of Cyprinus carpio variants in Cyprinidae.
Cyprinus carpio var. Guilin rice flower carp is a traditional domestic strain of common carp cultured in the paddy fields of Guilin mountain areas of Yunnan Province in China (Wang Citation1979; Jiang et al. Citation2014). To reveal evolutionary taxonomy and conserve the genetic resource, we analyzed the complete mitochondrial genome of C. carpio var. Guilin to identify their phylogenetic positions and intraspecific genetic variations.
Caudal fin samples of Cyprinus carpio var. Guilin were collected from Quanzhou County, Guilin City, Guangxi Province, China (E111°06′, N25°96′) (Laslett and Canback Citation2008). Total genomic DNA was extracted from the caudal fin tissues using DNA extraction kit (Tiangen, Beijing, China). Leftover DNA and specimen, with the tag name Cyprinus carpio var. Guilin, were deposited at the herbarium of Pearl River Fisheries Research Institute, Guangzhou, China. The genome was sequenced using Illumina HiSeq2500 platforms (San Diego, CA, USA) and the phylogenetic tree was constructed by MEGA6.0 software (University Park, PA, USA).
The complete mitochondrial genomes of C. carpio var. Guilin was 16,532 bp (MK291479) in length, which consisted of 22 tRNA genes, 13 protein-coding genes, 2 rRNA genes, and a D-loop region, which is consistent with other teleosts (Nardi et al. Citation2001; Wen et al. Citation2017). The overall nucleotide composition was 31.9% A, 24.9% T, 27.5% C, 15.7% G, with 56.8% AT, respectively.
Within 13 protein-coding genes, 12 genes started with the common initiation codon ATG, except COI began from GTG start codon. For stop codon, seven genes were terminated with TAA codon, three genes (DN2, ATPase 8 and ND3) with TAG codon, and three genes (COII, ND4, and Cytb) with incomplete stop codons (T–). Ten regions of gene overlapping ranged from 1 to 7 bp and ten intergenic spacer regions ranged from 1 to 33 bp. The results were the same as C. carpio (Chang et al. Citation1994).
12S rRNA (956 bp) and 16S rRNA (1,679 bp) were located between tRNA-Phe and tRNA-Leu and separated by tRNA-Val. A total of 22 tRNA genes varying from 66 to 75 bp were interspersed throughout the genome. The D-loop control region is 927 bp in length which is located between the tRNA-Pro and tRNA-Phe genes, as generally shown in most teleost mitochondrial genome (Chang et al. Citation1994; Yu and Wang Citation2017).
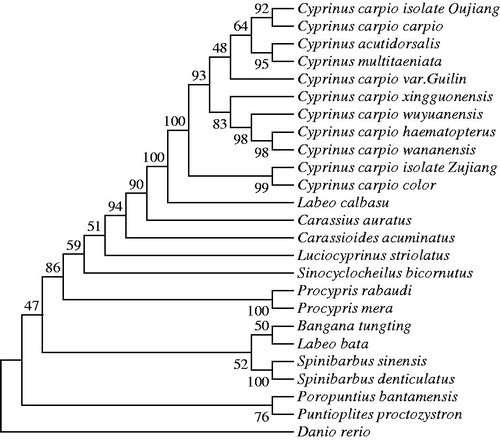
The phylogenetic tree was constructed on the basis of the complete mitochondrial genome sequences from Cyprinus carpio var. Guilin and other 24 species of Cyprinidae in the GenBank database. The phylogenetic tree showed that Cyprinus carpio var. Guilin is similar to C. carpio isolate Oujiang, C. carpio carpio, C. acutidorsalis, and C. multitaeniata within one cluster of Cyprinus. The species of C. carpio, C. acutidorsalis, and C. multitaeniata cannot be identified by the mitogenome sequences (). Also, it is much closer to Cyprinus carpio isolate Oujiang by BLAST analysis at web servers of the NCBI. Comparison between C. carpio var. Guilin and C. carpio isolate Oujiang (KP993136.1) indicated that they had 99.72% identity. A total of 47 variation loci including 44 transitions, two transversions, and one indel were detected.
Figure 1. The phylogenetic tree of 25 mitochondrial genomes with Danio rerio as an outgroup based on the neighbor-joining (NJ) and maximum likelihood (ML) analysis by Mega 7.0 program. The bootstrap values were calculated based on 1000 replications. The GenBank accession numbers of the sequences presented are as follows: Cyprinus carpio isolate Oujiang (KP993136.1), Cyprinus carpio carpio (JN105352.1), Cyprinus carpio haematopterus (JX188254.1), Cyprinus carpio wananensis (KF856964.1), Cyprinus carpio wuyuanensis (JN105357.1), Cyprinus carpio xingguonensis (JN105353.1), Cyprinus carpio isolate Zujiang (KP993137.1), Cyprinus carpio color (JX188253.1), Cyprinus acutidorsalis (KR869144.1), Cyprinus multitaeniata (KR869145.1), Carassius auratus (KJ874431.1), Labeo calbasu (JQ231113.1), Luciocyprinus striolatus (AP012525.1), Carassioides acuminatus (KX602324.1), Procypris rabaudi (EU082030.1), Procypris mera (JX316027.1), Bangana tungting (KJ737371.1), Spinibarbus sinensis (KF214722.1), Sinocyclocheilus bicornutus (KX528071.1), Spinibarbus denticulatus (AP013335.1), Poropuntius bantamensis (AP011352.1), Puntioplites proctozystron (AP011247.1), Labeo bata (AP011198.1), Danio rerio (AC024175.3).
Disclosure statement
The authors report that they have no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Chang YS, Huang FL, Lo TB. 1994. The complete nucleotide sequence and gene organization of carp (Cyprinus carpio) mitochondrial genome. J Mol Evol. 38:138–155.
- Jiang Y, Zhang S, Xu J, Feng J, Mahboob S, Al-Ghanim KA, Sun XW, Xu P. 2014. Comparative transcriptome analysis reveals the genetic basis of skin color variation in common carp. PLoS ONE. 9:e108200.
- Laslett D, Canback B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24: 172–175.
- Nardi F, Carapelli A, Fanciulli PP, Dallai R, Frati F. 2001. The complete mitochondrial DNA sequence of the basal hexapod Tetrodontophora bielanensis: evidence for heteroplasmy and tRNA translocations. Mol Biol Evol. 18: 1293–1304.
- Wang YK. 1979. Classification, distribution, origin and evolution of Cyprininae in China [J]. Acta Hydrobiol Sinica. 6:419–438. (in Chinese)
- Wen ZY, Xie BW, Qin CJ, Wang J, Yuan DY, Li R, Zou YC. 2017. The complete mitochondrial genome of a threatened loach (Beaufortia kweichowensis) and its phylogeny. Conservation Genet Resour. 9:565–568.
- Yu L, Wang G. 2017. Complete mitochondrial genome of Ctenopharyngodon idella var. Gold grass carp and its intraspecific comparison. Mitochondrial DNA A. 28: 372–374.
