Abstract
The complete chloroplast genome from Cardiocrinum cathayanum (Liliaceae), an important herbaceous perennial species of the Liliaceae, is determined in this study. We characterized its whole plastid genome sequence using the Illumina sequencing platform. The whole plastid genome of C. cathayanum was a typical quadripartite circular molecule with 152,502 bp in length, comprised of a large single copy region of 82,517 bp, a small single copy region of 17,311 bp, and two inverted repeat regions of 26,337 bp. A total of 136 genes, 84 protein-coding genes, 38 tRNA, and 8 rRNA genes were identified. The GC content in the chloroplast genome, LSC region, SSC region, and IR region were 42.5, 34.9, 30.8, and 42.5%, respectively. Phylogenetic analysis suggested that C. cathayanum is more closely related with Allium cepa, Allium prattii, Amana edulis, and Erythronium sibiricum, these species were clustered into a clade with high bootstrap support.
Cardiocrinum cathayanum is a species of Liliaceae, which is a long-lived, monocarpic, perennial herb mainly distributed in Qinling Mountains, Shaanxi Province, China, with distinct individual differences in plant height, the manner of flowering and floral characteristics. In addition, C. cathayanum was found to have important ornamental, edible, and medicinal value (Zhou Citation2015). However, the predicted suitable habitats of C. cathayanum have become smaller and more fragmented since the last glacial maximum. Therefore, this species needs to be better studied on species identification and molecular phylogenetic analysis as a conservation and exploitation target. In plants, chloroplast DNA sequence could provide valuable phylogenetic signals and in contrast to nuclear genomes, plant chloroplast genomes showed high copy numbers per cell and a much smaller size for complete sequencing (McNeal et al. Citation2006). In this study, we characterized the complete chloroplast genome sequence of C. cathayanum based on the Illumina pair-end sequencing data, which was an important way for future studies on population dynamics and further contribute to conservation strategy of the species.
Genomic DNA was extracted from fresh leaves of an individual of C. cathayanum collected from Qinling Mountains (N33.837361, E108.804111; the specimen (L-2018-102) was deposited at Northwest University), using the modified CTAB method (Doyle and Doyle Citation1987). Then, the DNAs were subjected to Illumina sample preparation, and pair-read sequencing was indexed by the Illumina Hiseq 2500 platform. In total, about 64,970 high-quality reads were obtained. The clean reads were assembled by the MIRA 4.0.2 program (Chevreux et al. Citation2004) and MITObim v1.7 software (Hahn et al. Citation2013). The chloroplast genomes were annotated using the program DOGMA (Wyman et al. Citation2004), coupled with manual corrections for start and stop codons. Finally, the chloroplast genome sequences were deposited in GenBank (accession numbers, KM104128).
Cardiocrinum cathayanum chloroplast complete genome was 152,511 bp in length, which comprise a large single copy (LSC) region of 82,517 bp and a small single copy (SSC) region of 17,311 bp, separated by two inverted repeat regions (IRs) of 26,337 bp. The circular genome contained 136 genes, including 84 protein-coding genes, 38 tRNA, and 8 rRNA genes. Most gene species occurred in a single copy, while 20 gene species occurred in double copies. The overall GC content of C. cathayanum chloroplast genome was 42.5%, while the corresponding values of LSC, SSC, and IR regions were 34.9, 30.8, and 42.5%, respectively.
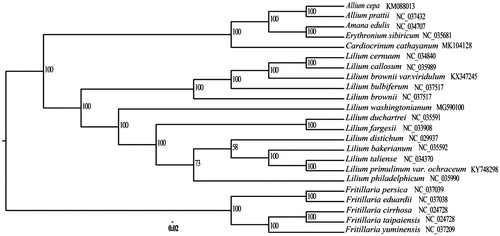
A total of 18 species from Lilium, Amana, Erythronium, and Allium were used to construct the phylogenetic tree with five Fritillaria species as outgroups. All of the 23 plastid sequences were aligned using the software MAFFT (Katoh and Standley Citation2013) with the default parameters. The phylogenetic analysis was conducted using the program RAxML (Stamatakis Citation2006) with 1000 bootstrap replicates (). The analysis suggested that C. cathayanum was more closely related with Allium cepa, Allium prattii, Amana edulis, and Erythronium sibiricum and these species were clustered into a clade with high bootstrap support. We believe that C. cathayanum chloroplast genome will provide valuable insight into conservation, utilization, and evolutionary histories for this important species.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Müller WE, Wetter T, Suhai S. 2004. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 14:1147–1159.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Boil Evol. 30:772–780.
- McNeal JR, Leebens-Mack JH, Arumuganathan K, Kuehl JV, Boore JL, DePamphilis CW. 2006. Using partial genomic fosmid libraries for sequencing complete organellar genomes. Biotechniques. 41:69–73.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zhou ZG. 2015. Research on breeding technology and seeds dispersal ability of Cardiocrinum cathayanum [Master dissertation]. Nanjing: Nanjing Forestry University.