Abstract
The complete mitochondrial (mt) genome of Pteropurpura falcatu was determined using genome walking techniques in this study. The total length of the mt genome sequence of P. falcatu was 15,319 bp, including 13 protein-coding genes, 22 transfer RNA genes, and 2 ribosomal RNA genes. The overall composition of the mitogenome was estimated to be 29.4% for A, 38.3% for T, 15.6% for C, and 16.7% for G, respectively, indicating that an A + T (67.7%)-rich feature occurs in the P. falcatu mitogenome. The phylogenetic relationships of 12 mollusc species were constructed based on the complete mtDNA sequences by the neighbor-joining method using MEGA 7.0 and DNAMAN 6.0 software.
The Pteropurpura falcatu belongs to the family Muricidae with very small distribution and could only be found in northern parts of Yellow Sea of China. The population of wild P. falcatu is small too, which results in difficulties to collect samples for the experiment. In addition, the research about P. falcatu is rare. In order to restore and protect the wild P. falcatu resources, it is necessary to carry out wild resource investigation and germplasm analysis. In this study, we report the complete sequence of mitochondrial genome for P. falcatu (GenBank accession no. MK348224). The findings will provide useful information for further studies on population genetics, phylogenetic construction, and other relevant studies in P. falcatu.
One P. falcatu individual was collected from Dalian Sea area, Liaoning Province, China (39°11′25″N, 124°47′25″E). The total genomic DNA was extracted from foot muscle by a modification of standard phenol-chloroform procedure. The complete mitogenome of P. falcatu was sequenced by primer walking. The gene annotation was performed following the methods described by Hao et al. (Citation2016) and Online software (GeneMarker, US; tRNAscan, US; ORFfinder, US). At present, the specimen is stored in the herbarium of Dalian Ocean University.
The total length of the hybrid of P. falcatu mitochondrial genome was 15,319 bp with the base composition of 29.4% A, 38.3% T, 15.6% C, and 16.47% G. It comprised of 2 ribosomal RNA genes, 13 protein-coding genes, and 22 transfer RNA genes. All the mitogenome genes were encoded on the heavy strand except for seven tRNA genes (tRNA-Met, tRNA-Tyr, tRNA-Cys, tRNA-Trp, tRNA-Glnp, tRNA-Gly, and tRNA-Glu).
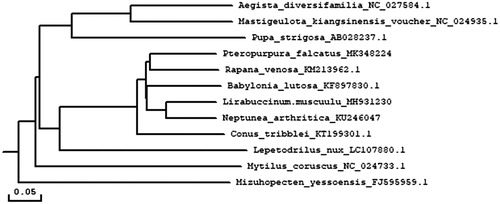
The phylogenetic analysis showed that the complete mitochondrial sequence of P. falcatu was phylogenetically closer to Rapana venosa by 78.09% bootstrap support and formed the Muricidae clade (). Our result was consistent with the previous researches, both in traditional morphological and molecular-based phylogeny studies. We expect that the present result can contribute to construct molecular identification of this species and be helpful to explore the phylogeny of Muricidae.
Figure 1. Consensus neighbor-joining tree based on the complete mitochondrial sequence of P. falcatu and other 11 mollusc. species. The phylogenetic tree was constructed using MEGA 7.0 and DNAMAN 6.0 software by the neighbor-joining method. The numbers at the tree nodes indicates the percentage of bootstrapping after 1000 replicates.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
Additional information
Funding
Reference
- Hao Z, Yang L, Zhan Y, Tian Y, Wang L, Mao J, Chang Y. 2016. The complete mitochondrial genome of Neptunea arthritica cumingii Crosse, (Gastropoda: Buccinidae). Mitochondr DNA B. 1:220–221.
