Abstract
The complete mitochondrial genome of hybrid F2 from Acanthopagrus schlegelii (♀) × Pagrus major (♂) was obtained using high-throughput sequencing technology. The circular genome was 16,648 bp in length, consisting of 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and a control region. Overall, nucleotide composition was A: 28.02%, T: 27.88%, C: 27.93%, G: 16.17%. Nine genes were encoded on the light strand and the remaining 28 genes were encoded on the heavy strand. All protein initiation codons are ATG except for COI that begins with GTG. The phylogenetic analysis suggested that hybrid F2 of A. schlegelii (♀) × P. major (♂) was closest to A. schlegelii. The newly described mitochondrial genome may provide valuable data for the genetic and taxonomic research on artificial hybrid seabream.
Both black seabream (Acanthopagrus schlegelii) and red seabream (Pagrus major) are important commercial species in the coastal waters of China, Japan, and Korea (Sadovy and Cornish Citation2000). Black seabreams have the advantage of delicious taste, low mortality, and low-temperature resistance whereas red seabream have the advantage of fast growth and good performance (Gwo et al. Citation2005). The hybrid F2 of A. schlegelii (♀) × P. major (♂) has significant hybridization advantages. Therefore, the complete mitogenome of hybrid F2 of A. schlegelii (♀) × P. major (♂) were sequenced to provide useful information for the genetic and taxonomic research on artificial hybrid seabream.
In this study, the sample was obtained by artificial hybridization from Fish Breeding Base of Marine Fisheries Research Institute of Jiangsu Province, Lvsi port, China (N 32°05′59.50″, E121°35′49.09″). The sample was numbered F219 and stored at the Marine Fisheries Research Institute of Jiangsu Province, Nantong, China. Total DNA was extracted with standard phenol-chloroform methods (Sambrook et al. Citation1989). The complete genome sequence of this sample (GenBank accession no. MK182268) is 16,648 bp in length which was sequenced by high-throughput sequencing technology (Illumina HiSeq 2000, USA). It encoded 37 genes totally containing 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, and a non-coding region (D-loop). All these genes were encoded on the H-strand, except for the ND6 subunit gene and eight tRNA genes (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, tRNAPro, and tRNAGlu) which were encoded on the L-strand. The total length of 13 protein-coding genes is 11,443 bp accounting for 68.74% of the complete genome. All the protein initiation codons are ATG, except for COI that starts with GTG. Seven PCGs (ND1, ND2, ATP6, COIII, ND4L, ND5, ND6) terminate with the stop codon TAA while the COI and ATP8 genes terminate with the stop codon AGG and TAG, respectively. The incomplete stop codon (T–) is widely used in the remaining four PCGs (COII, ND3, ND4, Cyt b). Overall base composition of the complete mitochondrial DNA is A: 28.02%, T: 27.88%, C: 27.93%, G: 16.17%.
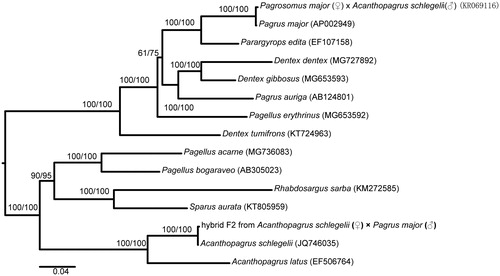
The phylogenetic trees were reconstructed based on the complete mitochondrial genome of the hybrid F2 and the available sequences of 14 Sparidae species, using maximum likelihood (ML) and neighbor-joining (NJ) methods in MEGA X (Kumar et al. Citation2018). The mitogenome sequences were aligned using Muscle in MEGA X and subsequently edited and trimmed. A total of 15,824 bp were used for phylogenetic analyses. The phylogenetic tree () showed that the hybrid F2 of A. schlegelii (♀) × P. major (♂) has a closer relationship with A. schlegelii (female parent) than the other species. In addition, the tree revealed two well-supported clades of Sparidae, indicating great mitochondrial divergence within the Sparidae (Orrell and Carpenter Citation2004; Zhu et al. Citation2016). This study improves our understanding of the inheritance of mitochondrial DNA in artificial hybrid offspring.
Figure 1. Phylogenetic tree based on mitochondrial genome nucleotide sequences of the hybrid F2 of Acanthopagrus schlegelii (♀) × Pagrus major (♂) and other 14 species of Sparidae reconstructed using the ML and NJ methods. Numbers above each branch represent ML and NJ bootstrap supports. GenBank accession number of each species is shown in parentheses.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Gwo JC, Chiu JY, Lin CY, Su Y, Yu SL. 2005. Spermatozoal ultrastructure of four sparidae fishes: Acanthopagrus berda, Acanthopagrus australis, Lagodon rhomboids and Archosargus probatocephus. Tissue Cell. 37:109–115.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Orrell TM, Carpenter KE. 2004. A phylogeny of the fish family Sparidae (porgies) inferred from mitochondrial sequence data. Mol Phylogenet Evol. 32:425–434.
- Sambrook J, Maniatis TE, Fritsch EF. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbour Laboratory Press; p. 1.
- Sadovy Y, Cornish AS. 2000. Reef fishes of Hong Kong. Hong Kong: Hong Kong University Press.
- Zhu F, Zhang Z, Chen S, Zhang Z, Xu J, Jia C, Yin S, Zhang G. 2016. The complete mitochondrial genome of the hybrid of Pagrus major (♀) × Acanthopagrus schlegelii (♂). Mitochondrial DNA A DNA Mapp Seq Anal. 27:2980–2981.
