Abstract
Dalbergia cochinchinensis is an tree species in Southeast Asia, its wood and wood products are incredibly valuable and are also of important medicinal value. In this study, its chloroplast genome was characterized using next generation Illumina pair-end and Pacbio sequencing dataset. The whole genome was 156,576 bp in length and contains a pair of 25,682 bp inverted repeat regions, which were separated by a large single copy region and a small single copy region of 85,886 and 19,326 bp in length, respectively. The cp genome contained 111 genes, including 77 protein-coding genes, 30 tRNAs and 4 rRNAs. A neighbor-joining phylogenetic analysis suggested D. cochinchinensis, which belonged to Dalbergieae, Fabaceae.
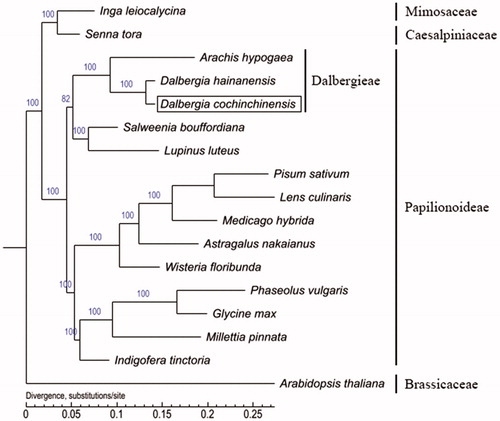
Phylogenetic analysis
Dalbergia cochinchinensis Pierre ex Laness is widely distributed in Thailand, Laos, Cambodia, and other Southeast Asian regions. It is a semi-deciduous or evergreen broad-leaved tree species in the Fabaceae family (leguminous) and takes more than 50 years to mature (Khorn Citation2002). D. cochinchinensis is used for high-grade rosewood furniture, handicraft, musical instruments, and traditional medicine. However, the natural resource of it was exhausted, which is caused by narrow and scattered distribution and long-time overexploitation (So et al. Citation2010). It is listed in the ‘CITES Appendix II’ and has been classified as an endangered plant listed in the category of key protected wild plants in the original country (CITES 2013). Some researches focused on biology (Fa and CTSP 2003), cultivation (Sovu et al. Citation2010), population diversity (Hartvig et al. Citation2018), wood chemical constituents (Liu et al. Citation2016) and medicinal research (Rasheeha et al. Citation2013). However, there has been no study about the whole chloroplast genome information of D. cochinchinensis until now. In this study, we reconstructed the complete chloroplast (cp) genome of D. cochinchinensis to provide the underlying information for genetic tree breeding and conservation studies.
Fresh leaves of D. cochinchinensis were collected from 2-year-old seedling-tree, which originated from Province Cha Choeng Sao, Thailand (13°21′36″N, 101°4′48″E), growing in rosewood national forest tree germplasm resources repository, Ledong District, Hainan Province, China (18°41′24″N, 108°47′24″E) and was harvested for cpDNA isolation using an improved extraction method (McPherson et al. Citation2013). High-throughput sequencing was carried out on the Illumina HiSeq 2000™ and PacBio RS II (Pacific Biosciences, California, USA) platform following the manufacturer’s protocol (Borgstrom et al. Citation2011). The chloroplast genes were annotated using an online DOGMA tool (Wyman et al. Citation2004). Phylogeny was constructed based on whole cpDNAs with 16 Fabaceae and Arabidopsis thaliana as outgroup species by the maximum-likelihood (ML) methods using PhyML 3.0 (http://www.atgc-montpellier.fr/phyml/) (Guindon et al. Citation2010).
The complete and annotated cp genome sequence of D. cochinchinensis had been submitted to GenBank with the accession number SRX4104984. There was a circular molecule 156,576 bp in length for the complete chloroplast genome of D. cochinchinensis, included a quadripartite structure that consists of a large single copy (LSC) region of 85,886 bp and a small single copy (SSC) region of 19,326 bp, separated by two inverted repeat (IR) regions of 25,682 bp. It encoded 111 genes, including 77 protein-coding genes, 30 tRNAs, and 4 rRNAs. Among these, nine genes contain a single intron while three genes possess two introns. Six PCGs, 7 tRNAs genes, and 4 rRNAs genes were duplicated in both IR regions. PCG ycf1 was a pseudogene in the IRa region. However, there still was a PCG (ycf1) in IRb and SSC region. The overall GC content was 36.08%.
The result indicated that D. cochinchinensis is clustered together with D. hainanensis and A. hypogaea, what was confirmed D. cochinchinensis was that it belonged to Dalbergia genera (). The complete cp genome information reported in this study would be useful for population genomic studies, conservation and tree breeding works on D. cochinchinensis as well as for phylogenetic studies within Leguminosae.
Acknowledgements
The authors are grateful to the opened raw genome data from public database. The authors thank Aiting Lin for the help of sequence analysis.
Disclosure statement
The authors declare no conflicts of interest and are responsible for the content.
Additional information
Funding
References
- Borgstrom E, Lundin S, Lundeberg J. 2011. Large scale library generation for high throughput sequencing. PLoS One. 6:e19119.
- CITES. Co P16 Prop. 60. 2013. Consideration of proposals for amendment of appendices I and II[C]. Bangkok (Thailand): Convention on International Trade in Endangered Species (CITES); p. 1–16
- Forestry A, CTSP 2003. Forest gene conservation strategy. Phnom Penh: Forestry Administration (FA) and Cambodia Tree Seed Project (CTSP), (Annex 2); p. B2-1–B2-9
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321.
- Hartvig I, So T, Changtragoon S, Tran HT, Bouamanivong S, Theilade I, Kjaer ED, Nielsen LR. 2018. Population genetic structure of the endemic rosewoods Dalbergia cochinchinensis and D. oliveri at a regional scale reflects the Indochinese landscape and life-history traits. Ecol Evol. 8:530–545.
- Khorn S. 2002. Distribution of selected tree species for gene conservation in Cambodia. Viet Nam forest trees. Hanoi: Forest Inventory and Planning Institute. Agricultural Publishing House: Annex; p. B2-1–B2-9.
- Liu RH, Li YY, Shao F, Chen LY, Wen XC, Zhang PZ, Huang HL. 2016. A new Chalcone from the heartwood of Dalbergia cochinchinensis. Chem Nat Compd. 52:405–408.
- McPherson H, van der Merwe M, Delaney SK, Edwards MA, Henry RJ, McIntosh E, Rymer PD, Milner ML, Siow J, Rossetto M. 2013. Capturing chloroplast variation for molecular ecology studies: a simple next generation sequencing approach applied to a rainforest tree. BMC Ecol. 13:8.
- Rasheeha N, Iftikhar H, Abdul T, Muhammad T, Moazur R, Sohail H, Shahid M, Abu BS, Mazhar I. 2013. Antimicrobial activity of the bioactive components of essential oils from Pakistani spices against Salmonella and other multidrug resistant bacteria [J]. BMC Complement Alternat Med. 13:265.
- So T, Theilade I, Dell B. 2010. Conservation and utilization of threatened hardwood species through reforestation – an example of Afzelia xylocarpa (Kruz.) Craib and Dalbergia cochinchinensis Pierre in Cambodia. Pacific Conserv Biol. 16:101–116.
- Sovu TM, SP, Odén PC, Xayvongsa L. 2010. Enrichment planting in a logged-over tropical mixed deciduous forest of Laos. J Forestry Res. 21:273–280.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.