Abstract
Apostasia shenzhenica is a terrestrial orchid, which belongs to Apostasioideae (Orchidaceae). Here, we determined the complete plastid genome of A. shenzhenica using the Illumina reads in an effort to provide genomic resources useful for its conservation and evolutionary biology study. It has a circular molecular length of 153,164 bp and contained 109 genes, including 75 protein-coding genes, 4 rRNA genes, and 30 tRNA genes. Phylogenetic analysis indicated that A. shenzhenica was closely related to A. wallichii and embedded in Apostasioideae.
Orchidaceae is one of the largest families of flowering plants consisting of five subfamilies. All of the wild orchids are protected by the International Trade Convention due to their Endangerment (Favre Citation1989; Chase et al. Citation2015). The Apostasia is one of the two genera that form subfamily Apostasioideae which was first diverged from Orchidaceae approximate 90 Ma (Givnish et al. Citation2015). The species Apostasia shenzhenica indigenous in South Guangdong in China is a terrestrial species with an actinomorphic perianth, unspecialized labellum, distinct stamens, and powdery pollen which distinguished from other Orchidaceae species (Chen and Liu Citation2011; Zhang et al. Citation2017). The genome of A. shenzhenica (total length 349 Mb) was assembled with high quality and inspired us to understand the origins and divergent evolution of orchids (Zhang et al. Citation2017). However, as a trademark of plant systematics and evolution, the plastid genome of A. shenzhenica has not been reported.
In order to assemble the plastid genome of A. shenzhenica, total compressed 3.4 G data of high-quality reads (NCBI accession # SRR5759388), which was released by the Apostasia genome sequencing project, were downloaded using the fastq-dump software https://ncbi.github.io/sra-tools/fastq-dump.html. The individual for sequencing was cultivated in the National Orchid Conservation Center of China and the sample was stored at NOCC herbarium in Shenzhen, China with the accession number ASH160606 (Zhang et al. Citation2017). These reads were then used to assemble the complete chloroplast genome with the plastid genomes of Calanthe triplicata (NC_024544.1) and Apostasia wallichii (LC_199394.1) as references. We performed the assembling and annotation using Geneious R10 Geneious R10 (https://www.geneious.com/) and adjusted the genes manually to make sure that they were maintained as open reading frames. IR boundaries for the draft plastome were confirmed by BLAST. Finally, we obtained a chloroplast genome of A. shenzhenica and submitted the whole genome to GenBank (MK370661).
The whole plastid genome of A. shenzhenica was 153,164 bp in length and contained two inverted repeat regions (IRA and IRB) of 27,510 bp, which is separated by a large single-copy (LSC) region of 86,167 bp and a small single-copy (SSC) region of 11,977 bp. The plastid genome of A. shenzhenica comprised 109 genes, including 75 protein-coding genes, 4 ribosomal RNA genes, and 30 transfer RNA genes. The overall GC content of A. shenzhenica chloroplast genome is 35.9% and the corresponding values in LSC, SSC, and IR regions are 32.7, 28.1, and 42.5%, respectively.
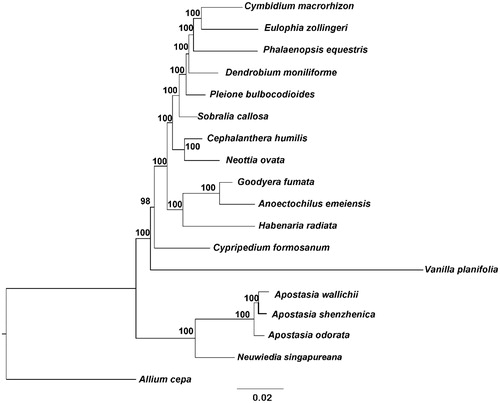
The phylogenetic tree was reconstructed based on whole plastid genome of 16 species representing all five orchid subfamilies and Allium cepa as outgroup. All protein-coding genes exported from Geneious were aligned with MAFFT (Katoh and Standley Citation2013) and subsequently adjusted manually in Bioedit v5.0.9 (Hall Citation1999). Aligned sequences were concatenated using SequenceMatrix (Vaidya, Lohman, and Meier Citation2011). A maximum-likelihood (ML) analysis of the plastome data was performed with IQtree (Nguyen et al. Citation2015) with 1000 ultrafast bootstrap (UFBoot) replicates (Minhet al. Citation2013; Chernomor et al. Citation2016). Phylogenetic analysis showed that A. shenzhenica was closely related to A. wallichii within Apostasioideae with strong support. The relationships of the five Orchidaceae subfamilies are well resolved based on sampled plastid genome in this study () .
Figure 1. Phylogenetic tree (maximum likelihood) based on the protein-coding genes of 16 orchids and Allium cepa. Numbers above branch are maximum parsimony bootstrap percentages. These accession numbers are as follows: Allium cepa (KM088013), Anoectochilus emeiensis (NC_033895), Apostasia odorata (KM244734), Apostasia wallichii (LC199394), Cephalanthera humilis (NC_030706), Cymbidium macrorhizon (NC_029713), Cypripedium formosanum (KJ501998), Dendrobium moniliforme (NC_035154), Eulophia zollingeri (NC_037212), Goodyera fumata (KJ501999), Habenaria radiate (KX871237), Neottia ovata (NC_030712), Neuwiedia singapureana (LC199503), Phalaenopsis equestris (NC_017609), Pleione bulbocodioides (NC_036342), Sobralia callosa (NC_028147) and Vanilla planifolia (KJ566306).
Authors’ contributions
S.S.W. and Z.J.L.conceived the study; Y.X.L and Z.H.L. obtained the molecular data; J.W.Z., Q.L.H., and Y.X.L. participated in data analysis; Y.X.L.and Z.H.L. drafted the manuscript; S.S.W., J.W.Z. and Z.J.L. revised the manuscript. All authors provided comments and final approval.
Acknowledgement
We are grateful to Shenzhen Key Laboratory for Orchid Conservation and Utilization, The National Orchid Conservation Centre of China, and The Orchid Conservation and Research Centre of Shenzhen for their support.
Disclosure statement
There are no conflicts of interest for all the authors.
References
- Chase MW, Cameron KM, Freudenstein JV, Pridgeon AM, Salazar G, Van den Berg C, Schuiteman A. 2015. An updated classification of Orchidaceae. Bot J Linn Soc. 177:151–174.
- Chen L, Liu Z. 2011. Apostasia shenzhenica, a new species of Apostasioideae (Orchidaceae) from China. Plant Sci J. 29:38–41.
- Chernomor O, von Haeseler A, Minh BQ. 2016. Terrace aware data structure for phylogenomic inference from supermatrices. Syst Biol. 65:997–1008.
- Favre DS. (1989). International Trade in Endangered Species: A Guide to CITES. Kluwer Academic Publishers, Dordrecht, Netherlands.
- Givnish TJ, Spalink D, Ames M, Lyon SP, Hunter SJ, Zuluaga A, Iles WJD, Clements MA, Arroyo MTK, Leebens-Mack J, et al. 2015. Orchid phylogenomics and multiple drivers of their extraordinary diversification. Proc R Soc B. 282:20151553.
- Hall TA. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic acids symposium series. 41:95–98.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Minh BQ, Nguyen MAT, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30:1188–1195.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Vaidya G, Lohman DJ, Meier R. 2011. SequenceMatrix: concatenation software for the fast assembly of multi‐gene datasets with character set and codon information. Cladistics. 27:171–180.
- Zhang G-Q, Liu K-W, Li Z, Lohaus R, Hsiao Y-Y, Niu S-C, Wang J-Y, Lin Y-C, Xu Q, Chen L-J, et al. 2017. The Apostasia genome and the evolution of orchids. Nature. 549:379.
