Abstract
A complete mitogenome of Acheilognathus chankaensis was assembled and analyzed in this study. The genome, with the length of 16,926 bp, contains 20 tRNAs, 13 coding genes, two rRNAs, and a D-loop region. The components of the genome are the same as available Acheilognathus mitogenomes in NCBI database, and GC (%) is also similar with other published mitogenomes in this genus. Phylogenetic analysis showed that A. chankaensis was genetically closed to A. macropterus. This complete mitogenome could contribute to the future phylogenetic and phylogeographic studies of genus Acheilognathus.
Acheilognathus chankaensis, known as Khanka bitterling, is a temperate freshwater fish belonging to the Acheilognathinae subfamily. It originates in the inland rivers and lakes and is found in China, Korea, and Russia. Acheilognathus chankaensis is benthopelagic and algophagous (Liu and Chang Citation2015) and have been consumed for a long history in China. Recently, due to its small size and nuptial coloration, A. chankaensis becomes a popular ornamental fish (Chen et al. Citation2009). Although it is a common fish in East Asia, there are quite a few genetic studies about it.
Herein, a newly assembled mitogenome of A. chankaensis (NCBI ID: MK380726) was characterized. Acheilognathus chankaensis (ID: y003) was collected from Yuan River, Hunan Province in China (28°87N, 112°34E) and preserved in 99% ethanol in Museum of Hunan Agricultural University. After DNA extraction and sequencing library construction, paired-end reads were sequenced using Illumina HiSeq (Illumina, San Diego, CA). Following Elbrecht et al. (Citation2015), the mitogenome of A. rhombeus (GenBank No. AP013342) was used to identify DNA sequences and extract all mitogenome reads from the A. chankaensis sequences using the discontinuous megablast algorithm BLASTn in the Geneious software package (max e-values 1e-20) (R11, Biomatters Ltd, Auckland, New Zealand). Extracted reads were assembled by mapping to the reference mitogenome of A. rhombeus. The first draft genome was manually checked and then used as a search query to identify additional reads in the Illumina sequences data. The mitogenome was annotated for Coding DNA Sequence (CDS), tRNA, and rRNA by comparison with all available Acheilognathus mitogenomes in GenBank (n = 14). The ORFs finder and Annotation (Vertebrate Mitochondrial) functions in Geneious were used to check the position, start, and stop codons and length of each CDS. After assembling the mitogenomes, multiple alignment (ClustalW) was utilized to compare differences among the mitogenomes in Acheilognathus by Geneious. MrBayes was employed to construct the phylogenetic tree with Rhodeus atremius as an outgroup, and the substitution model, rate variation, MCMC chain length, and burnin length were GTR, invgamma, 10,000,000, and 500,000, respectively.
The assembled genome contained 30,196 reads from the sequences databases, with good overall coverage (minimum coverage was 22 sequences, maximum coverage was 4242 sequences, average coverage was 319 (SD = 612.8). The total length of the genome was 16,926 bp, which included 20 tRNAs, 13 coding genes, two rRNAs, and a D-loop region. The base composition of the genome was A (29.9%), T (27.5%), C (26.0%), and G (16.6%).
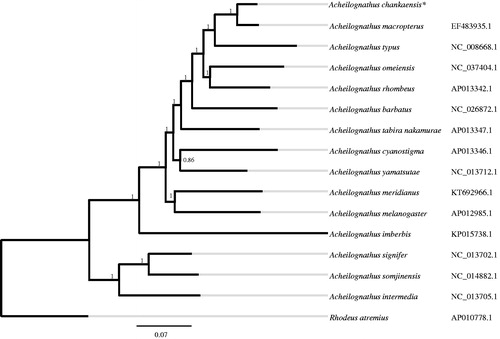
The full mitochondrial genome of A. chankaensis was slightly longer than that of other Acheilognathus species. The small difference between the species is primarily due to a ∼400 bp insertion in the D-loop region. Phylogenetic analysis showed that A. chankaensis clustered together with A. macropterus (), which was in accordance with their morphology (Cao et al. Citation2012). Furthermore, the genetic differentiations amongst Acheilognathus were relatively low (), and this would not surprise because the external morphology of this genus was very close to each other (Naseka and Bogutskaya Citation2004). The information of the mitogenome will be beneficial for future phylogenetic and phylogeographic studies of Acheilognathus species, particularly for A. chankaensis, and useful where there may be some confusion with specimen identification.
Figure 1. Bayesian phylogenetic tree based on the complete mitochondrial genome sequence of Acheilognathus chankaensis. The numbers along the branches were posterior probability. The asterisks after species names indicate newly determined mitochondrial genomes, and the codes followed the Latin names were NCBI access IDs for each mitogenomes.
Acknowledgements
We thank Jianbo Yu who collected the specimen.
Disclosure statement
There are no conflicts of interest for all the authors including the implementation of research experiments and writing this article.
Additional information
Funding
References
- Elbrecht V, Poettker L, John U, Leese F. 2015. The complete mitochondrial genome of the stonefly Dinocras cephalotes (Plecoptera, Perlidae). Mitochondrial DNA. 26:469–470.
- Liu F, Chang YL. 2015. Preliminary study on the biological characteristics of Acheilognathus chankaensis in Luohe river. Hebei Fisheries. 8:28–30.
- Naseka A, Bogutskaya N. 2004. Contribution to taxonomy and nomenclature of freshwater fishes of the Amur drainage area and the Far East (Pisces, Osteichthyes). Zoosyst Rossica. 12:279–290.
- Cao YH, Liao FC, Wu YA. (2012). Aquatic Fauna in Xiang River. Hunan: Hunan Science and Technology Press. p. 120–129.
- Chen L, Zhou CQ, Wang H, Qi WH, Liu GK. 2009. Morphometric characteristics, length-weight relationship and fullness index of Acheilognathus chankaensis. J China West Normal Univ (Nat Sci). 30:388–391.
