Abstract
Identification of invasive whitefly Bemisia tabaci cryptic species is best achieved by an integrated approach combining mating and genomic studies including complete mitochondrial DNA genomes (mitogenomes) characterization. We present the draft mitogenome of a 1920 historical B. tabaci specimen obtained using High Throughput Sequencing (HTS) technology. Our B. tabaci partial mtCOI gene best matched the B. tabaci ‘NW’ (New World) cryptic species (e.g. to AY057133; nucleotide identity: 99.85%). It has a circular genome of ca. 15,325bp with 13 protein-coding genes (PCGs), 22 tRNA’s, and two rRNA genes. A total of 43 amino acid residues differed between a Sanger-sequenced ‘NW’ mitogenome (AY521259.2) and this HTS-generated ‘NW’ mitogenome. High intra-specific amino acid conservation was observed across the partial 657 bp mitochondrial DNA COI (mtCOI) gene region used in species identification (i.e. from nucleotides 782 to 1438) when compared between both ‘NW’ (MK386668, AY521259.2) species.
Historical specimens are invaluable genomic resources to understand species habitat range (Tay et al. Citation2012, Tay, Elfekih, Polaszek, et al. Citation2017), species origins prior to global trade activities (Tay, Elfekih, Court, et al. Citation2017; Elfekih, Etter, et al. Citation2018), and can assist with identification of cryptic/morpho-species complexes (De Souza et al. Citation2016; Wash et al. Citation2019). The whitefly Bemisia tabaci is a cryptic species complex and species identification has relied on its partial mtCOI gene (De Barro et al. Citation2011); however, pseudogenes and PCR artefacts (Tay, Elfekih, Court, et al; Vyskočilová et al. Citation2018) associated with poor PCR markers (Elfekih, Tay, et al. Citation2018) represent significant challenges to molecular species identification. HTS approaches can effectively bypass these challenges by enabling rapid mitogenome characterization. The mitogenomes of one NW (via PCR/Sanger sequencing; Thao et al. Citation2004) and two ‘NW2’ (via HTS; De Marchi et al. Citation2018) species endemic in the New World have been reported; and, here, we report the first HTS-generated draft mitogenome of a historical B. tabaci ‘NW’ species (MK386668).
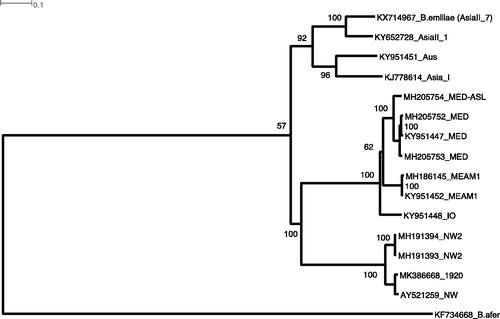
A single 4th instar ‘Bemisia tabaci’ puparium collected on October 1920 by Francis Watts from Barbados on the native tropic/sub-tropic plant host, peacock flower (Caesalpinia pulcherrima) (USNM dry collection, Q20358), was extracted following the methods of Tay, Elfekih, Court, et al. (Citation2017). We used Illumina Nextera-XT library preparation kit to prepare the genomic library using 1.5 ng of extracted gDNA as template. We used Geneious v11.1.5 to assemble the mitogenome against both the ‘NW’ (AY521259.2) and the ‘NW2’ (MH191393) mitogenomes, and Mitos (Bernt et al. Citation2013) for genome annotation, with fine-tuning carried out within Geneious. Gene orientations of the Barbados 1920 ‘NW’ mitogenome was conserved as for all B. tabaci cryptic species complex (e.g. Wang et al. Citation2012; Tay et al. Citation2016). We constructed a phylogenetic analysis involving concatenation of the 13 coding gene sequences using IQ-TREE webserver (Nguyen et al. Citation2015; Trifinopoulos et al. Citation2016) with 1000 bootstrap replications ‘Auto’ substation model selection. With the presence of pseudogenes and Sanger sequencing of potential PCR artefacts complicating species identification, HTS-generated B. tabaci cryptic species draft mitogenomes will serve as valuable genomic resources to better understand the evolutionary biology of these globally invasive agricultural insect pest species ().
Figure 1. Maximum Likelihood phylogenetic analyses of 15 Bemisia tabaci cryptic species mitogenomes and one B. afer mitogenome as an outgroup using IQTree with 1000 bootstrap replications involving 10,556 bp representing 13 concatenated coding gene aligned sequences with end trimming and gaps moved. The historical 1920 Barbados B. tabaci ‘NW’ species (MK386668) clustered with the Sanger sequenced NW (AY521259, Thao et al. Citation2004), and together with the two NW2 mitogenomes (De Marchi et al. Citation2018), formed a ‘New World’ clade with 100% bootstrap value for node support. Note that the AsiaII_1 mitogenome (KY652728) was available as an unpublished genome from GenBank (accessed 07-Jan-2019).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler P. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- De Barro PJ, Liu S-S, Boykin LM, Dinsdale AB. 2011. Bemisia tabaci: a statement of species status. Annu Rev Entomol. 56:1–19.
- De Marchi BR, Kinene T, Wainaina JM, Krause-Sakate R, Boykin L. 2018. Comparative transcriptome analysis reveals genetic diversity in the endosymbiont Hamiltonella between native and exotic populations of Bemisia tabaci from Brazil. PLoS One. 13:e0201411.
- De Souza BR, Tay WT, Czepak C, Elfekih S, Walsh TK. 2016. The complete mitochondrial DNA genome of a Chloridea (Heliothis) subflexa (Lepidoptera: Noctuidae) morpho-species. Mitochondrial DNA A DNA Mapp Seq Anal. 27:4532–4533.
- Elfekih S, Etter P, Tay WT, Fumagalli M, Gordon K, Johnson E, De Barro P. 2018. Genome-wide analyses of the Bemisia tabaci species complex reveal contrasting patterns of admixture and complex demographic histories. PLoS One. 13:e0190555.
- Elfekih S, Tay WT, Gordon K, Court LN, De Barro PJ. 2018b. Standardized molecular diagnostic tool for the identification of cryptic species within the Bemisia tabaci complex. Pest Manag Sci. 74:170–173.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Tay WT, Evans GA, Boykin LM, De Barro PJ. 2012. Will the real Bemisia tabaci please stand up? PLoS One. 7:e50550.
- Tay WT, Elfekih S, Court L, Gordon KH, De Barro PJ. 2016. Complete mitochondrial DNA genome of Bemisia tabaci cryptic pest species complex Asia I (Hemiptera: Aleyrodidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:972–973.
- Tay WT, Elfekih S, Polaszek A, Court LN, Evans GA, Gordon KH, De Barro PH. 2017. Novel molecular approach to define pest species status and tritrophic interactions from historical Bemisia specimens. Sci Rep. 7:429.
- Tay WT, Elfekih S, Court LN, Gordon KHJ, Delatte H, De Barro PJ. 2017. The trouble with MEAM2: Implications of pseudogenes on species delimitation in the globally invasive Bemisia tabaci (Hemiptera: Aleyrodidae) cryptic species complex. Genome Biol Evol. 9:2732–2738.
- Thao ML, Baumann L, Baumann P. 2004. Organization of the mitochondrial genomes of whiteflies, aphids, and psyllids (Hemiptera, Sternorrhyncha). BMC Evol Biol. 4:25.
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucl. Acids Res. 44: W232–W235.
- Vyskočilová S, Tay WT, van Brunschot S, Seal S, Colvin J. 2018. An integrative approach to discovering cryptic species within the Bemisia tabaci whitefly species complex. Sci Rep. 8:10886.
- Wash TK, Perera OP, Anderson CJ, Gordon KHJ, Czepak C, McGaughran A, Zwick A, Hackett DS, Tay WT. (2019). Mitochondrial DNA genome resources of five major Helicoverpa pest species from the Old and New Worlds (Lepidoptera: Noctuidae). Ecol Evol. 9:2933–2944.
- Wang XW, Zhao QY, Luan JB, Wang YJ, Yan GH, Liu SS. 2012. Analysis of a native whitefly transcriptome and its sequence divergence with two invasive whitefly species. BMC Genomics. 13:529.
