Abstract
In this study, we report the complete mitochondrial genome sequence of the Endangered Band-rumped Storm Petrel (Oceanodroma castro), a globally distributed seabird. The mitogenome is 17,023 bp in length and has a base composition of A (30.5%), T (24.0%), C (31.2%), and G (14.3%). Similar to other avifauna, it contains 13 protein-coding genes, two ribosomal RNA genes, 22 transfer RNA genes, and a control region, with arrangement and orientation identical to that of other seabirds. To our knowledge, this is the first complete mitochondrial genome sequenced within the family Hydrobatidae, or storm petrels, and will aid in taxonomic studies.
The Band-rumped Storm Petrel (Oceanodroma castro), although globally distributed, was recently listed under the Endangered Species Act in the Pacific region (USFWS 2015). Once widespread in the Hawaiian Islands, as evidenced by midden sites (Harrison Citation1990), its range is now limited (Olson and James Citation1982; Raine et al. Citation2017).
We sequenced the complete mitochondrial genome of O. castro (GenBank accession number MK170187). Blood, tissue, and feather samples from 25 individuals were collected on the islands of Kaua‘i, Hawai‘i, Maui, and O‘ahu. Samples from museum specimens are stored at the Bernice Pauahi Bishop Museum. DNA was individually extracted from blood, feather, or tissue samples using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA). The extracted DNA was quantified with the AccuClear™ Ultra High Sensitivity dsDNA Quantitation Kit (Biotium, Hayward, CA) using two rows of standards. Due to low DNA yield, whole genome amplification was performed on individual samples with the REPLI-g UltraFast Mini-kit (Qiagen, Valencia, CA). Equal quantities of DNA from 10 individuals were pooled by island population to a total of 1 μg per library, and two libraries were prepared for reduced representation sequencing using the ezRAD protocol (Toonen et al. Citation2013) version 2.0 (Knapp et al. Citation2016). The pooled libraries were digested with the frequent-cutter restriction enzyme DpnII from New England Biolabs® (Ipswich, MA), and fragments between 300 and 700 bp in length were prepared for sequencing on the Illumina®MiSeq using the Kapa Biosystems (Wilmington, MA) Hyper Prep kit. The samples were amplified to generate 1 μg of adapter-ligated DNA, then validated and quantified to ensure equal pooling on the Illumina flow cell, using a Bioanalyzer and qPCR. Quality control checks and sequencing on the MiSeq flow cell were performed by the Hawai‘i Institute of Marine Biology Genetics Core Facility for the pooled samples. Another 24 libraries from 24 individuals were prepared using the same protocol as described for pooled libraries, except samples were prepared individually and fragments were size selected between 150 and 350 bp in length for sequencing on the Illumina®HiSeq. Quality control checks, qPCR, and sequencing on the HiSeq flow cell were performed by Vincent J. Coates Genomics Sequencing Laboratory at the University of California, Berkeley for the individual samples.
We obtained 650,048,040 sequences. Reads were paired, then mapped to the mitogenome of Pterodroma brevirostris (Slack et al. Citation2006) using Geneious 10.2.6 (Biomatters, Newark, NJ). In total, 2,953,756 reads, or about 0.45% of reads, mapped to the mitochondrial genome, with coverage ranging from 279 × to 359,324 × per site (30,636 ± 61,578). Annotation of mitochondrial elements was carried out with DOGMA (Wyman et al. Citation2004) and MITOS (Bernt et al. Citation2013).
The O. castro mitogenome is 17,023 bp in length with a base composition of A (30.5%), T (24.0%), C (31.2%), and G (14.3%). The genes’ arrangement and orientation are identical to that of typical avian mtDNA (Gibb et al. Citation2006). Duplication was not detected in this study, in contrast to duplication of the control region observed in other storm petrels (Gibb et al. Citation2013).
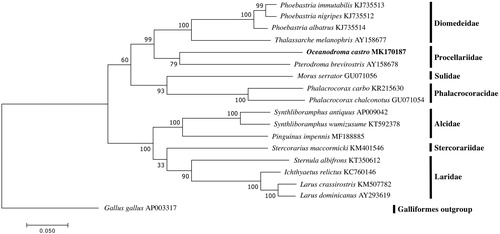
Figure 1. Placement of Oceanodroma castro among seabird families. Alignments, model tests, and maximum-likelihood analyses were performed using MEGA version 7. The 13 protein-coding mitochondrial gene sequences were translated into amino acid sequences, then aligned using ClustalW. Default settings were used with the following exception: the multiple alignment parameters were changed to a gap opening penalty of 3.0, and the gap extension penalty was set to 1.8. The amino acid substitution model was found to be JTT + G + F using the Akaike Information Criterion (AIC). Maximum-likelihood analysis of the amino acid sequences was run using the identified model, with bootstrap support values based on 1000 replicates. Gallus gallus was selected as the outgroup. The resulting tree shows similar relationships to previous studies (Nishibori et al. Citation2003; Yamamoto et al. Citation2005; Slack et al. Citation2006; Slack et al. Citation2007; Gibb et al. Citation2013; Lounsberry et al. Citation2015; Han et al. Citation2016; Eo and An Citation2016; Kim and Park Citation2016; Yang et al. Citation2016; Thomas et al. Citation2017; Zhang et al. Citation2017).
Collection site
Samples were collected from 19°38′N, -155°32′E.
Acknowledgements
We appreciate the invaluable assistance in sample collection provided by Nicole Galase, Molly Hageman, Dr. David Hyrenbach, and Dr. Andre Raine.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Eo SH, An J. 2016. The complete mitochondrial genome sequence of Japanese murrelet (Aves: Alcidae) and its phylogenetic position in Charadriiformes. Mitochondrial DNA A. 27:4574–4575.
- Gibb GC, Kardailsky O, Kimball RT, Braun EL, Penny D. 2006. Mitochondrial genomes and avian phylogeny: complex characters and resolvability without explosive radiations. Mol Biol Evol. 24:269–280.
- Gibb GC, Kennedy M, Penny D. 2013. Beyond phylogeny: pelecaniform and ciconiiform birds, and long-term niche stability. Mol Phylogenet Evol. 68:229–238.
- Han YD, Baek YS, Kim JH, Choi HG, Kim S. 2016. Complete mitochondrial genome of the South Polar Skua Stercorarius maccormicki (Charadriiformes, Stercorariidae) in Antarctica. Mitochondrial DNA A. 27:1783–1784.
- Harrison CS. 1990. Seabirds of Hawaii: natural history and conservation. Ithaca (NY): Cornell University Press.
- Kim JY, Park YC. 2016. The complete mitogenome of the Black-tailed gull Larus crassirostris (Charadriiformes: Laridae). Mitochondrial DNA A. 27:1885–1886.
- Knapp IS, Puritz JB, Bird CE, Whitney JL, Sudek M, Forsman ZH, Toonen RJ. 2016. ezRAD-an accessible next-generation RAD sequencing protocol suitable for non-model organisms v3.1. Protocols.io. Life Sciences Protocol Repository.
- Lounsberry ZT, Brown SK, Collins PW, Henry RW, Newsome SD, Sacks BN. 2015. Next-generation sequencing workflow for assembly of nonmodel mitogenomes exemplified with North Pacific albatrosses (Phoebastria spp.). Mol Ecol Resour. 15:893–902.
- Nishibori M, Hanazono M, Yamamoto Y, Tsudzuki M, Yasue H. 2003. Complete nucleotide sequence of mitochondrial DNA in White Leghorn and White Plymouth Rock chickens. Animal Sci J. 74:437–439.
- Olson SL, James HF. 1982. Fossil birds from the Hawaiian Islands: evidence for wholesale extinction by man before western contact. Science. 217:633–635.
- Raine AF, Holmes ND, Travers M, Cooper BA, Day RH. 2017. Declining population trends of Hawaiian Petrel and Newell's Shearwater on the island of Kaua‘i, Hawaii, USA. The Condor. 119:405–415.
- Slack KE, Delsuc F, Mclenachan PA, Arnason U, Penny D. 2007. Resolving the root of the avian mitogenomic tree by breaking up long branches. Mol Phylogenet Evol. 42:1–3.
- Slack KE, Jones CM, Ando T, Harrison GL, Fordyce RE, Arnason U, Penny D. 2006. Early penguin fossils, plus mitochondrial genomes, calibrate avian evolution. Mol Biol Evol. 23:1144–1155.
- Thomas JE, Carvalho GR, Haile J, Martin MD, Castruita JAS, Niemann J, Sinding M-HS, Sandoval-Velasco M, Rawlence NJ, Fuller E, et al. 2017. Anʻ Aukward’Tale: a genetic approach to discover the whereabouts of the Last Great Auks. Genes. 8:164.
- Toonen RJ, Puritz JB, Forsman ZH, Whitney JL, Fernandez-Silva I, Andrews KR, Bird CE. 2013. ezRAD: a simplified method for genomic genotyping in non-model organisms. PeerJ. 1:e203.
- United States Fish and Wildlife Service. [USFWS]. 2015. Endangered and Threatened Wildlife and Plants; Endangered a Status for 49 Species From the Hawaiian Islands; Proposed Rule. Federal Register: National Archives and Records Administration. 80.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yamamoto Y, Kakizawa R, Yamagishi S. 2005. Mitochondrial Genome Project of Endangered Birds in Japan: 1. Ancient Murrelet, Synthliboramphus antiquus. J Yamashina Inst Ornithol. 37:20–29.
- Yang C, Wang QX, Huang Y, Xiao H. 2016. Complete mitochondrial genome of Relict Gull, Larus relictus (Charadriiformes: Laridae). Mitochondrial DNA A. 27:411–412.
- Zhang L, Zhang M, He S. 2017. The complete mitochondrial genome of great cormorant, Phalacrocorax carbo (Phalacrocorax, Phalacrocoracidae). Mitochondrial DNA A. 28:1–2.
