Abstract
Potentilla fragarioides var. major Maxim. distributed in East Asia has been discussed about its scientific name. To clarify its phylogenetic position, we presented first complete chloroplast genome of P. fragarioides var. major which is 156,440 bp long and has four subregions: 85,784 bp of large single copy (LSC) and 18,616 bp of small single copy (SSC) regions are separated by 26,020 bp of inverted repeat (IR) regions including 129 genes (84 coding genes, eight rRNAs, and 37 tRNAs). The overall GC content of the chloroplast genome is 36.9% and those in the LSC, SSC, and IR regions are 34.8%, 30.7%, and 42.7%, respectively. Phylogenetic trees show that P. fragarioides var. major is sister to P. freyniana with high bootstrap values, suggesting further phylogenetic and morphological researches about the two species.
Potentilla is a neighbour genus of Fragaria containing strawberry species (Fragaria x ananassa) of which genomic studies have been conducted intensively (Shulaev et al. Citation2011; Hirakawa et al. Citation2014; Bai et al. Citation2017; Cheng et al. Citation2017; Han et al. Citation2018). In contrast, smaller genomic resources have been accumulated in Potentilla genus; three complete and several partial chloroplast genomes (Ferrarini et al. Citation2013; Zhang et al. Citation2017) including those of Duchesnea.
Potentilla fragarioides var. major Maxim. is one of Potentilla species distributed in East Asia. It does not develop creeping stems and has pinnately compound leaves (Lee et al. Citation2007). Because of these characteristics, it was considered Potentilla sprengeliana Lehm. (Naruhashi Citation2003); but later P. sprengeliana was treated as illegal name (Soják Citation2008). Li et al. regarded P. fragarioides var. major as a synonym of P. fragarioides L. (Li et al. Citation2003), indicating additional researches are required for clarifying this taxonomical argument. As a first step, we completed its chloroplast genome.
Total DNA of P. fragarioides var. major collected in Korea (Voucher in InfoBoss Cyber Herbarium (IN); K-I. Heo, IB-00574) was extracted from fresh leaves using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Genome sequencing was performed using HiSeq2000 at Macrogen Inc., Korea, and de novo assembly and confirmation were done using Velvet 1.2.10 (Zerbino and Birney Citation2008), SOAPGapCloser 1.12 (Zhao et al. Citation2011), BWA 0.7.17 (Li H 2013), and SAMtools 1.9 (Li H et al. Citation2009). Geneious R11 11.0.5 (Biomatters Ltd, Auckland, New Zealand) was used for chloroplast genome annotation based on Potentilla centigrana chloroplast complete genome (MK209637).
The chloroplast genome of P. fragarioides var. major (Genbank accession is MK227179) is 156,440 bp and has four sub regions: 85,784 bp of large single copy (LSC) and 18,616 bp of small single copy (SSC) regions are separated by 26,020 bp of inverted repeat (IR). It contains 129 genes (84 protein-coding genes, eight rRNAs, and 37 tRNAs); 17 genes (six protein-coding genes, four rRNAs, and seven tRNAs) are duplicated in IR regions. The overall GC content is 36.9% and those in the LSC, SSC, and IR regions are 34.8%, 30.7%, and 42.7%, respectively.
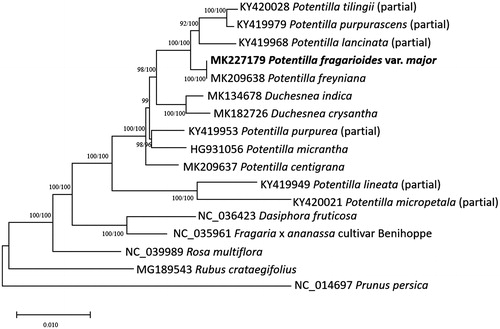
Ten partial or complete Potentilla, two Duchesnea, and five Rosaceae chloroplast genomes were used for constructing neighbor joining (bootstrap repeat is 10,000) and maximum likelihood trees (bootstrap repeat is 1,000) using MEGA X (Kumar et al. Citation2018) based on sequence alignment using whole chloroplast genomes using MAFFT 7.388 (Katoh and Standley Citation2013). Phylogenetic trees show that P. fragarioides var. major is clustered with Potentiila freyniana with high bootstrap support (), identifying nine single nucleotide polymorphisms (SNPs) and 48 insertions and deletions (INDELs) between two Potentilla species. It seems that both species are very close to each other in the aspect of molecular phylogeny, whereas morphological characters of radical leaves of the two species are different (Heo et al., under revision). Additional researches are required for understanding phylogenetic and taxonomical relationships of two species.
Figure 1. Neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenetic trees of seventeen Rosaceae partial or complete chloroplast genomes: Potentilla stolonifera (MK227179, in this study), Potentilla freyniana (MK209638), Potentilla centigrana (MK209637), Duchesnea chrysantha (MK182726), Duchesnea indica (MK134678), Potentilla tilingii (KY420028; partial genome), Potentilla lancinata (KY419968; partial genome), Potentilla purpurea (KY419953; partial genome), Potentila purpurascens (KY419979; partial genome), Potentilla micropetala (KY420021; partial genome), Potentilla micrantha (HG931056), Potentilla lineata (KY419949; partial genome), Dasiphora fruticosa (NC_036423), Fragaria x ananassa cultivar Benihoppe (NC_035961), Rubus crataegifolius (MG189543), Rosa multiflora (NC_039989), and Prunus persica (NC_014697). The numbers above branches indicate bootstrap support values of maximum likelihood and neighbor joining phylogenetic trees, respectively.
Disclosure statement
The authors declare that they have no competing interests.
Additional information
Funding
References
- Bai L-J, Ye Y-T, Chen Q, Tang H-R. 2017. The complete chloroplast genome sequence of the white strawberry Fragaria pentaphylla. Conserv Genet Resour. 9:659–661.
- Cheng H, Li J, Zhang H, Cai B, Gao Z, Qiao Y, Mi L. 2017. The complete chloroplast genome sequence of strawberry (Fragaria × ananassa Duch.) and comparison with related species of Rosaceae. PeerJ. 5:e3919.
- Ferrarini M, Moretto M, Ward JA, Šurbanovski N, Stevanović V, Giongo L, Viola R, Cavalieri D, Velasco R, Cestaro A, Sargent DJ. 2013. An evaluation of the PacBio RS platform for sequencing and de novo assembly of a chloroplast genome. BMC Genom. 14:670
- Han Y, Wu HB, Liu Y. 2018. The complete chloroplast genome sequence of Fragaria orientalis (Rosales: Rosaceae). Mitochondrial DNA B. 3:127–128.
- Heo K-I, Kim Y, Park* J, Lee* S. (In Submission). Taxonomic studies of the tribe Potentilleae (Rosaceae) in Korea. Korean J. Plant Res.
- Hirakawa H, Shirasawa K, Kosugi S, Tashiro K, Nakayama S, Yamada M, Kohara M, Watanabe A, Kishida Y, Fujishiro T, et al. 2014. Dissection of the octoploid strawberry genome by deep sequencing of the genomes of Fragaria species. DNA Res. 21:169–181.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Lee ST, Kim K-J, Lee B-Y, Choi B-H, Park J-H, Yang J-Y. 2007. Rosoideae. In: Park C-W, editor. The Genera of Vascular Plants of Korea. Seoul: Academic Publishing. 1482 p. ISBN. 745696222:543-567.
- Li C, Ikeda H, Ohba H, Zhengyi W, Raven P, Hong D. 2003. Potentilla L. Flora China. 9:291–328.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Naruhashi N. 2003. Rosoideae. In Iwatsuki K, Ohba H, Boufford DE, editors. Flora of Japan. Vol. IIb. Tokyo: Kodansha.
- Shulaev V, Sargent DJ, Crowhurst RN, Mockler TC, Folkerts O, Delcher AL, Jaiswal P, Mockaitis K, Liston A, Mane SP, et al. 2011. The genome of woodland strawberry (Fragaria vesca). Nat Genet. 43:109
- Soják J. 2008. Notes on Potentilla XXI. A new division of the tribe Potentilleae (Rosaceae) and notes on generic delimitations. Botanische Jahrbücher. 127:349–358.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhang SD, Jin JJ, Chen SY, Chase MW, Soltis DE, Li HT, Yang JB, Li DZ, Yi TS. 2017. Diversification of Rosaceae since the Late Cretaceous based on plastid phylogenomics. New Phytol. 214:1355–1367.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinform. 12:S2.
