Abstract
Pseudorimula is a vent-endemic genus found at some hydrothermal vents in West Pacific, North Atlantic, and Indian oceans. Here, we report a nearly complete mitogenome of Pseudorimula sp. (Vetigastropoda, Lepetodriloidea, Lepetodrilidae), which is collected from Tiancheng vent field on the Southwest Indian Ridge. The genome is 16,682 in length, including 13 complete protein-coding genes, two rRNA genes, and 22 tRNA genes. All genes are arranged in the same order as that in the genus Lepetodrilus, revealing a potential conserved gene arrangement in the family Lepetodrilidae. The phylogenetic analysis shows that the Pseudorimula+Lepetodrilus clade is well supported.
Deep-sea hydrothermal vents are characterized by extreme environmental gradients, which support vent fauna of unusually high biomass/abundance (Ramirez-Llodra et al. Citation2007). Pseudorimula is a genus of the family Lepetodrilidae (Desbruyères et al. Citation2006), which represents one of the most abundant and diverse family at deep-sea hydrothermal vent fields (Johnson et al. Citation2008). Studies based on the mitogenome of Lepetodrilus, the type genus of this family, revealed a novel gene arrangement in Vetigastropoda, and located this genus as sister to Granata and Haliotis (Lee et al. Citation2016; Nakajima et al. Citation2016). Psudorimula was once placed in Clypeosectidae of Fissurelloidea based on the shell, gill, and some anatomic characters (McLean Citation1989), but then transferred to Lepetodrilidae of Lepetodriloidea according to some other morphological characters (such as protoconch and radula) and COI fragments (Bouchet et al. Citation2005; Kano Citation2007). More robust genetic evidences are needed to resolve its phylogeny and biogeographic history; however, genetic information of Pseudorimula is largely unknown. Here, we describe the mitogenome of Pseudorimula sp. to better understand the phylogenetic location and the gene arrangement of the family.
During the Chinese cruise DY35 in December 2014, individuals of slit limpets were collected from Tiancheng vent field (63°32′E, 27°57′S, 2760 m depth) using the manned submersible Jiaolong, and then deposited in the Repository of the Second Institute of Oceanography, MNR, Hangzhou, China. The specimens were identified as Pseudorimula sp. based on the morphological characters and the partial COI sequence (Zhou et al. Citation2018). Genomic DNA was extracted with a Qiagen DNeasy Blood & Tissue Kit (69504) and then used for library construction. The resulting library was sequenced on an Illumina HiSeq4000 platform (Illumina Inc., San Diego, CA, USA). About 10 G clean data were de novo assembled using CLC Genomics Workbench v8.0 (Qiagen, CA, USA). A contig with a length of 16,682 bp was considered as the mitogenome (GenBank accession number: MK404176), on which protein-coding genes (PCGs), ribosomal RNA (rRNA) genes, and transfer RNA (tRNA) genes were annotated with MITOS (Bernt et al. Citation2013). The boundaries of PCGs were manually adjusted by comparison with published gastropod mitogenomes. Alignments of the amino acid sequences of the 13 PCGs were carried out with the MUSCLE algorithm (Edgar Citation2004). A maximum likelihood (ML) analysis based on the concatenated amino acid sequences was conducted in RAxML (Stamatakis Citation2014).
The nearly complete mitogenome of Pseudorimula sp. contains 13 PCGs, two rRNA genes, and 22 tRNA genes. All PCGs start with ATG, except for ND1, ND4, and ATP8, which use GTG as the start codon; most PCGs end with TAA codon, with the exception of COX3, ND2, and ND3, which terminate with TAG codon.
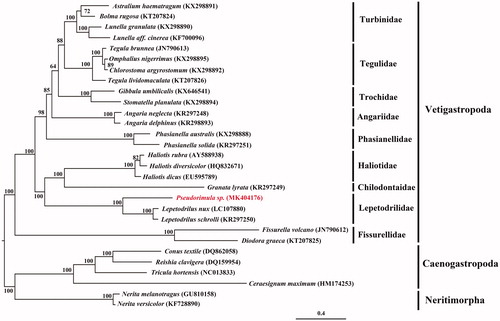
The gene order is identical to that in Lepetodrilus species (Nakajima et al. Citation2016), showing a conserved gene arrangement in this family. Phylogeny inference with maximum likelihood () robustly support that Pseudorimula is grouped with Lepetodrilus, forming a sister group to Granata and Haliotis within the Vetigastropoda.
Figure 1. Phylogenetic location of Pseudorimula sp. in the Vetigastropoda based on the maximum likelihood (ML) tree using the concatenated amino acid (AA) sequences of 13 PCGs of 29 species. ML bootstrap value is indicated at each node. Taxon groups are shown by vertical lines. Caenogastropoda and Neritimorpha species serve as the outgroup.
Acknowledgments
The authors thank all the scientists and crew on the R/V “Xiangyanghong 9”, and the submersible Jiaolong team for their help in the specimens’ collection during the expedition DY35; Chong Chen for his help with the specimens' identification.
Disclosure statement
The authors declare no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Bouchet P, Rocroi JP, Fr’yda J, Hausdorf B, Ponder WF, Valdés Á, &, Warén A, 2005. Classification and nomenclator of gastropod families. Malacologia. 47:1–397.
- Desbruyères D, Segonzac M, Bright M. 2006. Handbook of deep-sea hydrothermal vent. Second completely revised edition. Denisia 18:544 pp.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797.
- Johnson S, Warén A, Vrijenhoek RC. 2008. DNA barcoding of Lepetodrilus limpets reveals cryptic species. J Shellfish Res. 27:43–51.
- Kano Y. 2007. Vetigastropod phylogeny and a new concept of Seguenzioidea: independent evolution of copulatory organs in the deep-sea habitats. Zool Scr. 37:1–21.
- Lee H, Samadi S, Puillandre N, Tsai MH, Dai CF, Chen WJ. 2016. Eight new mitogenomes for exploring the phylogeny and classification of Vetigastropoda. J Mollus Stud. 82:534–541.
- McLean JH. 1989. New slit-limpets (Scissurellacea and Fissurellacea) from hydrothermal vents. Part 1. Systematic Descriptions and Comparisons Based on Shell and Radular Characters. Contributions in Science, Natural History Museum of Los Angeles County. 407:1–29.
- Nakajima Y, Shinzato C, Khalturina M, Nakamura M, Watanabe H, Satoh N, Mitarai S. 2016. The mitochondrial genome sequence of a deep-sea, hydrothermal vent limpet, Lepetodrilus nux, presents a novel vetigastropod gene arrangement. Mar Genom. 28:121–126.
- Ramirez-Llodra E, Shank TM, German CR. 2007. Biodiversity and biogeography of hydrothermal vent species thirty years of discovery and investigations. Oceanography. 20: 30–41.
- Stamatakis A. 2014. RaxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Zhou Y, Zhang D, Zhang R, Liu Z, Tao C, Lu B, Sun D, Xu P, Lin R, Wang J, Wang C. 2018. Characterization of vent fauna at three hydrothermal vent fields on the Southwest Indian Ridge: implications for biogeography and interannual dynamics on ultraslow-spreading ridges. Deep-Sea Res Pt I. 137:1–12.
