Abstract
From this study, the complete mitochondrial genome of Cicadella viridis was sequenced. Based on annotation, the mitogenome is circular, with 15,891 bp in length, containing 13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes, and one AT-rich region. There is 10,899 bp for the total length of 13 PCGs, which is encoding 3622 amino acids. All protein-coding genes used ATN as initiation codon. All protein coding genes used TAN as stop codon, except COII and COIII using incomplete T as stop codon. The length of LSU and SSU of rRNA gene are 1206 bp and 736 bp, respectively. Taxonomy of Cicadellidae was revised with phylogenetic analyses among mitogenome sequences from 20 closely related species.
The subfamily Cicadellinae belongs to the family Cicadellidae of order Hemiptera. The known species of this group are herbivorous, and some species are important pests on the fruit trees. The species of Cicadellinae are widely distributed throughout all major geographic regions of animal, especially in the tropical and subtropical regions. There are 26 genera of Cicadellinae in China. Species Cicadella viridis is one widespread species, with cylindrical body shape and 6.2–10.8 mm in body length (Yang et al. Citation2017).
A male adult of C. viridis was selected as specimen. It was collected from Kuankuoshui Reserve, Guizhou, China, in August 2018. Total DNA was extracted from entire body without abdomen. Qiagen DNeasy kit (Qiagen, Venlo, the Netherlands) was applied with the instruction of manufacture. Male genitalia is deposited in the Institute of Entomology, Guizhou University, Guiyang, China, for further study. Library was prepared and sequenced for 150 bp paired-end reads using HiSeqXten platform at Sangon Biotech (Shanghai, China). The reads was assembled and annotated using Geneious v9.1.5 (Aucland, New Zealand), with reference sequences from Homalodisca coagulata (accession number AY875213) and Bothrogonia ferruginea (accession number KU167550). According to the comparison results by Geneious, 37 typical invertebrate mitochondrial genes (13 PCGs, 22 tRNAs, and 2 rRNAs) and the A + T-rich region (D-loop) were identified. Annotated sequence of C. viridis mitogenome was submitted to GenBank with accession number MK335936.
The circular mitogenome of C. viridis is 15,891 bp in size. The A + T content of the C. viridis mitochondrial genome is 78.1%, which is within the range reported for hemipteran mitogenomes (Zhang et al. Citation2014). There is 10,899 bp for the total length of 13 PCGs, which is encoding 3622 amino acids. All protein-coding genes used ATN as initiation codon. All protein-coding genes used TAN as stop codon, except COII and COIII using incomplete T as stop codon. All tRNA genes have the conserved triple nucleotides recognizing corresponding codon. The length of LSU and SSU of rRNA gene are 1206 bp and 736 bp, respectively.
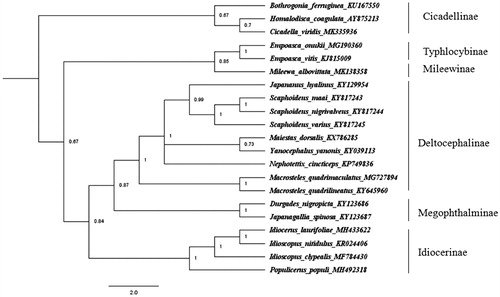
We used nucleotides sequences of 13 PCGs of C. viridis and 20 reference sequences from other species in family Cicadellidae species to construct phylogenetic relationship. Bayesian inference (BI) method was used through MrBayes 3.2.0 (Ronquist et al. Citation2012) under the GTR + G + I model and run for 10,000,000 generations with a sampling frequency of 100 generations. The results showed that C. viridis and other two species of subfamily Cicadellinae were clustered into one clade, which was separated from other subfamilies (). Mitochondrial gene sequences are important in solving the phylogenetic relationships. Therefore, the molecular data obtained in this study provided future resolution of the classification of family Cicadellidae.
Figure 1. Phylogenetic tree was constructed using MrBayes 3.2.0 (Ronquist et al. Citation2012) under the GTR + G + I model and run for 100,000 generations with a sampling frequency of 100 generations.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Ronquist K, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic Inference and model choice across a large model space. Syst Biol. 61:539–542.
- Yang MF, Meng ZH, Li ZZ. 2017. Hemiptera: Cicadellidae (II): Cicadellinae. Fauna Sinica: Insecta, vol. 67. Beijing (China): Science Press; p. 637.
- Zhang B, Ma C, Edwards O, Fuller S, Kang L. 2014. The mitochondrial genome of the Russian wheat aphid Diuraphis noxia: Large repetitive sequences between trnE and trnF in aphids. Gene. 533:253–260.
