Abstract
Anisodus tanguticus is endemic to the Qinghai-Tibetan Plateau, but its wild populations are shrinking roughly, and need urgent conservation. Here, we report the complete sequence of the chloroplast genome of A. tanguticus. The genome was 155,764 bp in length with 129 genes comprising 84 protein-coding genes, 37 tRNA genes, and eight rRNA genes. The overall GC content of A. tanguticus chloroplast genome was is 37.9%. Phylogenomic analysis suggested that A. tanguticus forms a monophyletic group with Hyoscyamus which shows closed relationship with the clade of Przewalskia and Scopolia.
Anisodus tanguticus (Maxim.) Pascher, belonging to Solanaceae, is endemic to the Qinghai-Tibetan Plateau in China (Wu and Raven Citation1994). As an important traditional Chinese medicine, it contains high levels of tropane alkaloids, scopolamine and scopolamine, which mainly affect the parasympathetic nervous system and can be used as an anticholinergic drug for acute glomerulonephritis, rheumatoid arthritis, hemorrhagic necrotic enteritis, eclampsia, pulmonary oedema, and circulatory shock (Yang Citation1991). However, the populations of A. tanguticus is beginning to dwindle due not only to the effect of excessive collection as a medicinal species (Zheng et al. Citation2007) but also to the low germination rate of the seeds (He and Jia Citation2009). Therefore, it has been listed as a critically endangered plant in the list of wild plants under state key protection in China (http://rep.iplant.cn/prot/Anisodus%20tanguticus). In recent years, most researches concerning A. tanguticus mainly have been focused on its chemical composition (Guo et al. Citation2015), pharmacological activity (Sun and Yang Citation2010), genetic diversity (Zheng et al. Citation2007), and functional gene identification (Liu et al. Citation2005) and less is known about the chloroplast gene of A. tanguticus. In the present study, we report the completed chloroplast genome of A. tanguticus based on the next-generation sequencing method. To our knowledge, this is the first completed chloroplast genome of Anisodus genus. The chloroplast genome will contribute to molecular phylogeny, genetic improvement, conservation, and sustainable management for this threatened species.
A wild individual of A. tanguticus was sampled from Qinghai, China (Voucher specimen: Zhang2018050, HNWP) and its genomic DNA was isolated from the fresh leaves with the modified CTAB method. DNA sample was randomly fragmented to construct paired-end (PE) libraries according to the Illumina preparation manual (San Diego, CA, USA). Genome sequences were screened out and assembled with SPAdes (Bankevich et al. Citation2012). Annotation was performed with CpGAVAS (Liu et al. Citation2012). The annotated genomic sequence had been submitted to GenBank with the accession number MF593117.
The complete chloroplast genome of A. tanguticus has a total length of 155,764 bp which composed of two inverted repeats (IR, 25,882 bp) separated by a large single copy region (LSC, 86,514 bp) and a small single copy region (SSC, 17,486 bp). The GC content of its chloroplast DNA is 37.9% (LSC, 35.6%; IR, 42.9%; SSC, 31.9%). The chloroplast genome was predicted to encode 129 genes, of which, 84 were protein-coding genes, 37 were distinct tRNA genes, and eight were rRNA genes. A total of 17 genes were duplicated in the IR regions including seven tRNA, four rRNA, and six protein-coding genes.
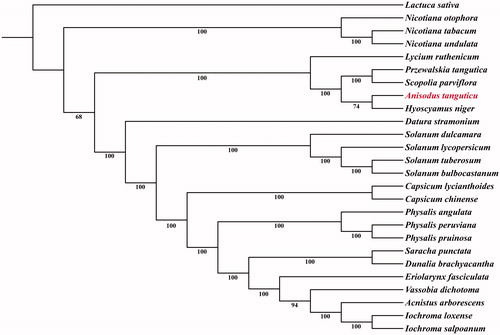
Phylogenetic analysis was performed based on complete chloroplast genomes of A. tanguticus and 24 other species from Solanaceae. Meanwhile, Lactuca sativa (Asteraceae) was used as an outgroup. Sequences were aligned using MAFFT v7.0 (http://mafft.cbrc.jp/alignment/server/) (Kazutaka and Standley Citation2013), and Gblocks (Castresana Citation2000) was employed to remove ambiguously aligned sites. A maximum likelihood (ML) analysis was conducted on RAxML-HPC2 on XSEDE based on the GTR + G + I nucleotide substitution model as recommended by jModelTest2 with 1000 replications.
The phylogenetic tree showed that the A. tanguticus forms a clade with Hyoscyamus niger which shows closed relationship with the clade of Przewalskia and Scopolia (). We expect that the chloroplast genome of A. tanguticus will be a valuable resource for future studies on conservation genetics, taxonomy, and phylogeny involving this particular species.
Disclosure statement
No potential conflict of interest was reported by the authors.
Figure 1. Maximum likelihood phylogenetic tree based on 26 complete chloroplast genome sequences. The number on each node indicates the bootstrap value. Accession numbers: Acnistus arborescens KU306735.1; Anisodus tanguticus MF539117; Capsicum chinense KU041709.1; Capsicum lycianthoides KP274856.1; Datura stramonium JN654342.1; Dunalia brachyacantha KP308151.1; Eriolarynx fasciculata KU306938.1; Hyoscyamus niger KF248009.1; Iochroma loxense KP296185.1; Iochroma salpoanum KU315119.1; Lactuca sativa AP007232; Lycium ruthenicum MG976805.1; Nicotiana otophora KU051626.1; Nicotiana tabacum Z00044.2; Nicotiana undulata JN563929.1; Physalis angulata MH019241.1; Physalis peruviana MH019242.1; Physalis pruinosa MH019243.1; Przewalskia tangutica KY352315.1; Saracha punctata KP280050.1; Solanum bulbocastanum DQ347958.1; Solanum dulcamara KY863443.1; Solanum lycopersicum HG975525.1; Scopolia parviflora KU900232.1; Solanum tuberosum DQ386163.2; Vassobia dichotoma KP294521.1.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J Comput Biol. 19:455–477.
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552.
- Guo H, Wu XL, Wang AL, Luo XW, Ma YJ, Zhou M. 2015. Separation and detection of tropane alkaloids in Anisodus tanguticus by capillary electrophoresis-electrochemiluminescence. New J Chem. 39:8922–8927.
- He T, Jia JF. 2009. Breaking dormancy in seeds of Anisodus tanguticus: an endangered medicinal herb of high altitude in the Qinghai-Tibet Plateau. Seed Sci Technol. 37:229–231.
- Kazutaka K, Standley DM. 2013. MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Liu C, Shi LC, Zhu YJ, Chen HM, Zhang JH, Lin XH, Guan XJ. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715.
- Liu T, Zhu P, Cheng K-D, Meng C, He H-X. 2005. Molecular cloning, expression and characterization of hyoscyamine 6β-hydroxylase from hairy roots of Anisodus tanguticus. Planta Med. 71:249–253.
- Sun K, Yang LM. 2010. Advance in pharmacological and clinical research of anisodamine. World Clin Drugs. 31:182–186.
- Wu ZY, Raven PH. 1994. Flora of China: vol. 17. Verbenaceae through Solanaceae. Beijing (China): Science Press, and St. Louis (MO): Missouri Botanical Garden Press.
- Yang YC. 1991. Flora Tebitan medicine. Xininng (China): Qinghai People's Publishing House.
- Zheng W, Wang LY, Meng L, Liu JQ. 2007. Genetic variation in the endangered Anisodus tanguticus (Solanaceae), an alpine perennial endemic to the Qinghai-Tibetan Plateau. Genetica. 132:123–129.
