Abstract
Xueduo yak (Bos grunniens) is one of the native yak genetic resources of China. Here, the complete mitochondrial genome of the Xueduo yak was first sequenced in this study. The total length of this mitochondrial genome is 16,323 bp, with the base composition of 33.72% A, 27.28% T, 25.80% C, and 13.20% G. The genome has 37 genes, including 13 protein-coding genes, 22 tRNA genes, and two ribosomal RNA genes. The phylogenetic analysis showed that Xueduo yak is closely related to Qinghai Plateau yak and Maiwa yak. This complete mitochondrial genome sequence of Xueduo yak would be useful for further understanding the evolution and conservation of Xueduo yak.
Yak is one of the most important domestic animals in Qinghai-Tibetan Plateau and in adjacent high-altitude regions (Wiener et al. Citation2003). There are approximately 5 million yaks domestic reside in the Qinghai Province, which accounts for 40% of the Chinese domestic yak population (Shi et al. Citation2016). They provide meat, milk, wool, transportation, and home heating for people living at high altitudes. Xueduo yak is a special yak genetic resource of Qinghai Province, China, which characterized by the better growth performance and higher disease resistance. Mitochondrial genome was considered as one of the most useful molecular tools in evolutionary and phylogenetic studies (Wang et al. Citation2011). However, less information about the complete mitochondrial genome of the Xueduo yak is available, which is necessary for conservation and utilization of the species. This study firstly reported complete mitochondrial DNA sequence of Xueduo yak and compared with other breeds.
The specimen of Xueduo yak was collected from Huangnan Tibetan Autonomous Prefecture, Qinghai Province, China (N34°44′8.14″, E101°36′53.15″). Muscle tissues were preserved in Key Laboratory of Yak Breeding Engineering of Gansu Province at −80 °C. Total genomic DNA was extracted from the skeletal muscle tissue using the animal tissue DNA isolation kit (ZDGSY, Beijing, China) according to the manufacturer’s instructions. DNA was stored at −20 °C. Six pairs of primers were used to amplify mitochondrial DNA by polymerase chain reaction, and the sequencing results were assembled using DNAStar 5.01 software.
We obtained the complete mitochondrial genome of Xueduo yak (GenBank Accession No. MK124956) in this study. Whole mitochondrial genome sequence has a circular genome of 16,323 bp, containing 13 protein-coding genes, two ribosomal RNA genes, 22 transfer RNA genes, and one non-coding control region (D-Loop region). The gene order and composition of the mitochondrial genome of Xueduo yak was similar to that of other yak species (Guo et al. Citation2016). The light strand (L-strand) was predicted to contain one protein-coding gene (ND6) and eight tRNA (Gln, Ala, Asn, Cys, Try, Glu, Pro, and Ser). The overall base composition of the genome is (33.72% A; 27.28% T; 25.80% C, and 13.20% G) with an overall A + T content of 61.00%.
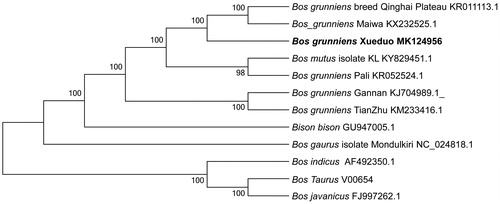
To ascertain the phylogenetic placement of Xueduo yak, a neighbour-joining (NJ) phylogenetic tree was constructed with MEGA6 (Tamura et al. Citation2013) using the coding sequences of 11 other bovine species. According to phylogenetic tree analysis, Xueduo yak has a closer genetic relationship with Qinghai Plateau yak and Maiwa yak (). In summary, the complete mitochondrial genome sequence of Xueduo yak can be further used for phylogenetic reconstruction and conservation strategies of this species.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Guo X, Pei J, Bao P, Chu M, Wu X, Ding X, Yan P. 2016. The complete mitochondrial genome of the Qinghai Plateau yak Bos grunniens (Cetartiodactyla: Bovidae). Mitochondrial DNA A. 27:2889–2890.
- Shi Q, Guo Y, Engelhardt SC, Weladji RB, Zhou Y, Long M, Meng X. 2016. Endangered wild yak (Bos grunniens) in the Tibetan plateau and adjacent regions: population size, distribution, conservation perspectives and its relation to the domestic subspecies. J Nat Conserv. 32:35–43.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wang Z, Yonezawa T, Liu B, Ma T, Shen X, Su J, Guo S, Hasegawa M, Liu J. 2011. Domestication relaxed selective constraints on the yak mitochondrial genome. Mol Biol Evol. 28:1553–1556.
- Wiener G, Han J, Long R. 2003. The Yak, 2nd ed. Bangkok (Thailand): Regional Office for Asia and the Pacific Food and Agriculture Organization of the United Nations.