Abstract
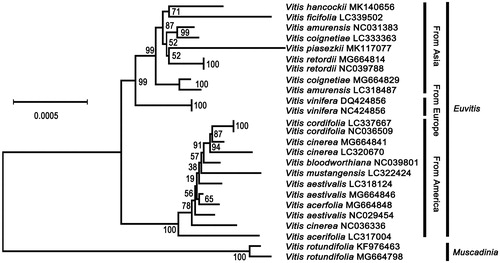
In this study, the complete chloroplast genome of Vitis hancockii was assembled for the first time. This genome was 161,205 bp in length, containing a large single copy region (89,397 bp) and a small single copy region (19,100 bp), seperated by two inverted repeat regions. The genome harbored 133 genes, including 88 protein-coding genes, 37 tRNA genes, and eight rRNA genes. Phylogenetic tree analysis showed that V. hancocikk was the closest related to Vitis ficifolia.
Vitis hancockii is wild oriental species, which mainly occur in the subtropical zones of China, such as Jiangxi, Zhejiang, Hubei, Anhui, Fujian, and Guizhou. This species highly resistant to powdery mildew (Wang et al. Citation1995), and mainly used as resistant germplasm for breeding new species. At present, the certain phylogenetic position of V. hancockii is uncertain, and there is no complete chloroplast genome sequence of any V. hancockii in the NCBI GeneBank database. We aimed to assemble the complete chloroplast genome of V. hancockii.
The fresh leaves of V. hancockii were obtained from Grape Germplasm and Breeding Research Vineyard of Shanghai Jiao Tong University, 800 Dongchuan Road, Minhang District, Shanghai, China (121°25′34.13″E; 31°01′11.15″N). DNA was extracted from fresh leaves using CTAB method (Qu et al. Citation1996) and stored at −80 °C in the Viticulture and Enology Center of Shanghai Jiao Tong University. Paired-end (150 bp) reads were sequenced by HiSeq X sequencing platform (Illumina, CA, USA). After discarding the low-quality data, 3.92 Gb clean data were used to assemble the complete chloroplast genomic using SOAPdenovo v2.04 (Luo et al. Citation2012) and MITObim v1.8. (Hahn et al. Citation2013). Chloroplast genome sequence of Vitis vinifira (DQ424856) was used as reference. DOGMA package was used to predict coding sequences (CDS), ribosomal RNA (rRNA), and transfer RNA (tRNA).
The complete chloroplast genome of V. hancockii (GeneBank: MK140656) was 161,205 bp in length. This genome included a large single copy region (LSC; 89,397 bp) and a small single copy region (SSC; 19,100 bp), separated by inverted repeat region A (IRA; 26,354 bp) and inverted repeat region B (IRB; 26,354 bp). The genome contained 133 annotated genes, including 88 protein-coding genes (PCGs), 37 tRNA genes, and eight rRNA genes.
To confirm the phylogenetic position of V. hancockii within Vitis, V. hancockii chloroplast sequence was aligned with those of other 24 Vitis species using MAFFT version 7 online page (Katoh and Standley Citation2013). Neighbour-joining phylogenetic tree () was constructed using MEGA X with 1000 bootstrap replicated. Phylogenetic tree indicated that all the 25 Vitis species were separated into Euvitis subgenus and Muscadinia subgenus. Vitis from America belonged to different branches of evolution compared to species from Europe and Asia. Vitis hancockii and Vitis ficifolia were most closely related to other Euvitis.
Disclosure statement
There are no conflicts of interest for all the authors including the implementation of research experiments and writing this article.
Additional information
Funding
References
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 13:e129–e129.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 4:772–780.
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 1:18
- Qu X, Lu J, Lamikanra O. 1996. Genetic diversity in muscadine and American bunch grapes based on randomly amplified polymorphic DNA (RAPD) analysis. J Am Soc Hortic Sci. 6:1020–1023.
- Wang Y, Liu Y, He P, Cheng J, Lamikanra O, Lu J. 1995. Evaluation of foliar resistance to Uncinula necator in Chinese wild Vitis species. Vitis. 3:159–164.