Abstract
Camellia japonica L. usually blooms red flower in winter in Jeju island, Korea, considered a species which should not be planted in the garden. Here, we completed chloroplast genome of C. japonica isolated in Seogwang-ri, Jeju island for comparing that isolated in Wimi-ri, Jeju island. Its length is 156,971 bp long and has four subregions: 86,673 bp of large single copy (LSC) and 18,394 bp of small single copy (SSC) regions are separated by 25,952 bp of inverted repeat (IR) regions including 135 genes (91 protein-coding genes, eight rRNAs, and 36 tRNAs). Its overall GC content is 37.3% and those in the LSC, SSC, and IR regions are 35.3%, 30.5%, and 43.0%, respectively. One insertion and one deletion are identified between chloroplast genomes of Soyeonpyeong island and Seogwang-ri, even though both places are far enough (>450 km). Twenty-five single nucleotide polymorphisms (SNPs) are identified within two Jeju island chloroplast genomes, presenting that C. japonica tree in Wimi-ri is genetically far from the two chloroplast genomes. Trees also show that genetic diversity of C. japonica is lower than that of C. sinensis.
Camellia japonica L. usually blooms beautiful red flowers in winter in Jeju island, Republic of Korea, called as rose of winter. Because of this red colour, Jeju people associate it with important person’ wrongful death, leading them consider that camellia should not be planted in the garden.
Camellia japonica chloroplast genome sampled in Wimi-ri, Jeju island was successfully assembled (Kim et al. Citation2017). Camellia trees in Wimi-ri are originated from tree bought by married women around 100 years ago as a wind break forest, indicating the possibility that Wimi-ri might not be a natural habitat so that genetic diversity of C. japonica in Jeju island will be valuable.
Total DNA of C. japonica collected in Seogwang-ri, Andeok-myeon, Seogwipo city (N33°18′18.54″, E126°17′29.53″; Voucher in InfoBoss Cyber Herbarium (IN); Y. Kim, IB-00582) was extracted from fresh leaves using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Genome was sequenced using HiSeqX at Macrogen Inc., Korea, and de novo assembly and confirmation were conducted with Velvet 1.2.10 (Zerbino and Birney Citation2008), SOAPGapCloser 1.12 (Zhao et al. Citation2011), BWA 0.7.17 (Li Citation2013), and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.0.5 (Biomatters Ltd, Auckland, New Zealand) was used for annotation based on C. japonica chloroplast genome (MK353210; Jongsun et al. Citation2019).
The C. japonica chloroplast genome in Seogwang-ri (Genbank accession is MK353211) is 156,971 bp long (GC ratio is 37.3%) and has four subregions: 86,673 bp of large single copy (LSC; 35.3%) and 18,394 bp of small single copy (SSC; 30.5%) regions are separated by 25,952 bp of inverted repeat (IR; 43.0%) regions. It contains 135 genes (91 protein-coding genes, eight rRNAs, and 36 tRNAs); 19 genes (eight protein-coding genes, four rRNAs, and seven tRNAs) are duplicated in IR regions.
Based on alignment of three C. japonica chloroplast genomes using MAFFT 7.388 (Katoh and Standley Citation2013), one insertion and one deletion on Seogwang-ri chloroplast genome are identified against Soyeonpyeong chloroplast genome; while, 25 single nucleotide polymorphisms (SNPs) are identified between Wimi-ri and Seogwang-ri inside Jeju (Jongsun et al. Citation2019). Considering straight-line distances between Soyeonpyeong and Seogwang-ri (>450 km) and between Seogwang-ri and Wimi-ri (∼30 km inside Jeju island), sequence variations on chloroplast genomes identified here is different from what we usually expected.
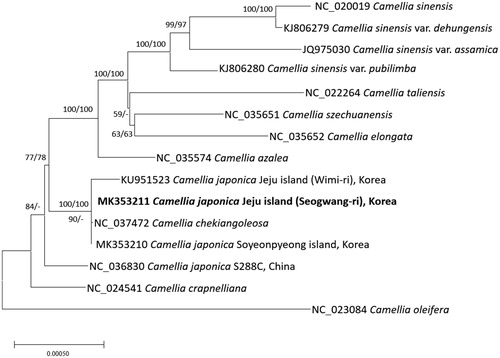
Fifteen chloroplast genomes of Camellia including four C. japonica chloroplast genomes were used for constructing neighbour-joining (bootstrap repeat is 10,000) and maximum likelihood phylogenic trees (bootstrap repeat is 1,000) using MEGA X (Kumar et al. Citation2018) after aligning whole chloroplast genomes using MAFFT 7.388 (Katoh and Standley Citation2013). Phylogenetic trees display that Wimi-ri chloroplast is distant from those of Soyeonpyeong and Seogwang-ri even in the same clade (); however, Chinese C. japonica is placed outside of the clade (). It indicates that additional chloroplast genomes sampled from Jeju island are needed to know origin of Wimi-ri’s camellia tree. In addition, four Camellia sienesis chloroplast genomes are more divergent than those of C. japonica (), presenting that C. japonica chloroplast genomes contain low sequence variations.
Figure 1. Neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenetic trees of 15 Camellia chloroplast genomes: Camellia japonica (MK353211 in this study, MK353210, KU951523, and NC_036830), Camellila chekiangoleosa (NC_037472), Camellia crapnelliana (NC_024541), Camellia oleifera (NC_024541), Camellia azalea (NC_035574), Camellia sinensis (NC_024541), Camellia sienesis var. pubilimba (KJ806280), Camellia sienesis var. assamica (JQ975030), Camellia sienesis var. dehungensis (KJ806279), Camellia szechuanensis (NC_035651), Camellia tallensis (NC_022264), Camellia elongata (NC_035652), and Camellia oleifera (NC_023084). The numbers above branches indicate bootstrap support values of maximum likelihood and neighbor joining phylogenetic trees, respectively.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Jongsun P, Yongsung K, Hong X, Yu JO, Kyung MH, Jaeyoung K. 2019. The complete chloroplast genome of common camellia tree, Camellia japonica L. (Theaceae), adapted to cold environment in Korea. Mitochondrial DNA Part B. 4(1):1038–1040. doi:10.1080/23802359.2019.1580164.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kim S-H, Cho CH, Yang M, Kim S-C. 2017. The complete chloroplast genome sequence of the Japanese Camellia (Camellia japonica L.). Mitochondrial DNA B. 2:583–584.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Preprint arXiv. 13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinform. 12:S2.
