Abstract
Here, we reported the complete mitogenome of Saccharina cultivar ‘Dongfang No.6’. This mitogenome had a circular mapping organization with the length of 37,657 bp and contained 66 genes including 35 protein-encoding genes, three rRNAs, 25 tRNAs, and three open reading frames (ORFs). The overall AT content was 64.70% exhibiting a high AT richness. The gene content and gene order were consistent with those reported Saccharina species and cultivars. Phylogenetic analysis showed phylogenetic relationship among Chinese Saccharina cultivars and indicated that ‘Dongfang No.6’ had a closer relationship with Saccharina japonica which strongly supported its female source.
Saccharina (Laminariales, Phaeophyceae) is an economically important brown macroalga (Kain Citation1979) which has contributed significantly to Chinese mariculture. ‘Dongfang No.6’, an intraspecific hybrid of Saccharina japonica, was high in yield and had a high stress tolerance (Li et al. Citation2016) which has been approved by Chinese Approving Committee of Aquacultural Elite Varieties and Stock Seeds (GS-02-004-2013). However, its information on genetics is limited. Here, we characterized the complete mitogenome of ‘Dongfang No.6’ and constructed phylogenetic analysis to provide new molecular data for the genetics study of this excellent Saccharina cultivar.
‘Dongfang No.6’ specimen (specimen number: 2016070089) was collected from Yantai, Shandong Province, China (37°52′N, 121°39′E) and stored at −80 °C for DNAs isolation. The homologous PCR amplification method and data processing were performed described by Zhang et al. (Citation2011).
The ‘Dongfang No.6’ mitogenome comprised a circular molecule of 37,657 bp (GenBank accession number MG712776), with a nucleotide composition of 28.41% A (10,699), 14.72%C (5544), 20.58% G (7750), and 36.29% T (13,664) and had an overall AT content of 64.70%. Cumulative GC-skew and AT-skew analysis reflected a slight bias toward G and T on H-strand. This mitogenome encoded three rRNAs (23S, 16S, and 5S), 25 tRNAs, 35 protein-encoding genes, and three ORFs (orf41, orf130, and orf377). The protein-encoding regions were 29,007 bp in length accounting for 77.03% of the whole mitogenome. With the exception of rpl2, rpl16, rps3, rps19, tatC, and ORF130, 60 genes were encoded on H-strand. The universal genetic codes were used and all start codons were ATG. Of the 35 protein-encoding genes and three ORFs, 26 (68.42%) terminated with TAA codon, higher than that for TAG (8, 21.05%) and TGA (4, 10.53%). The conserved gene cluster (rps8-rpl6-rps2-rps4) in the reported Saccharina mitogenomes was also found here. Moreover, all tRNA genes were provided with standard clover-leaf secondary structures. Gene content and gene order showed a high level of conservation with those reported Saccharina mitogenomes (Yotsukura et al. Citation2010; Zhang et al. Citation2013, Citation2017, Citation2018).
A comparison of the complete ‘Dongfang No.6’ mitogenome with that of S. japonica (Accession number: AP011493) showed they possessed the identical sequences indicating the high conservatism.
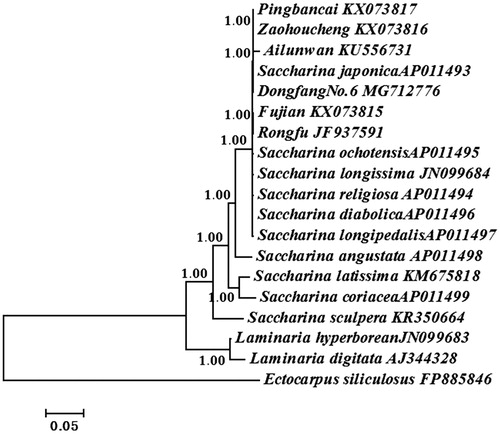
Bayesian analysis based on the whole mitogenomes of the 19 Saccharina and Laminaria algae was conducted to reconstruct the phylogeny (). Ectocarpus siliculosus served as an out-group. Phylogenetic analyses showed that all algae were divided into two branches: Saccharina and Laminaria. All Chinese cultivars including ‘Dongfang No.6’ belonged to the Saccharina lineage. ‘Dongfang No.6’ firstly clustered with S. japonica supporting its genetic origin. The phylogenetic tree in this work also provided more information for the current understanding of genetic relationships among Chinese Saccharina cultivars.
Disclosure statement
There are no conflicts of interest for all the authors including the implementation of research experiments and writing this article.
Additional information
Funding
References
- Kain JM. 1979. A view on the genus Laminaria. Oceanogr Mar Biol Annu Rev. 17:101–161.
- Li X, Zhang Z, Qu S, Liang G, Zhao N, Sun J, Song S, Cao Z, Li X, Pan J, et al. 2016. Breeding of an intraspecific kelp hybrid Dongfang no.6 (Saccharina japonica, Phaeophyceae, Laminariales) for suitable processing products and evaluation of its culture performance. J Appl Phycol. 28:439–447.
- Yotsukura N, Shimizu T, Katayama T, Druehl LD. 2010. Mitochondrial DNA sequence variation of four Saccharina species (Laminariales, Phaeophyceae) growing in Japan. J Appl Phycol. 22:243–251.
- Zhang J, Li N, Liu N, Liu T. 2017. The complete mitogenome of “Zaohoucheng”, an important Saccharina japonica cultivar in China. Mitochondrial DNA B Resour. 2:656–657.
- Zhang J, Liu N, Liu T. 2018. The complete mitogenome of “Fujian”: an important Saccharina japonica cultivar of South China. Mitochondrial DNA B Resour. 3:588–589.
- Zhang J, Li N, Zhang ZF, Liu T. 2011. Structure analysis of the complete mitochondrial genome in cultivation variety “Rongfu”. J Ocean Univ China. 10:351–356.
- Zhang J, Wang XM, Liu C, Jin YM, Liu T. 2013. The complete mitochondrial genomes of two brown algae (Laminariales, Phaeophyceae) and phylogenetic analysis within Laminaria. J Appl Phycol. 25:1247–1253.