Abstract
The complete mitochondrial genome sequence of Cylicocylus ashworthi was determined for the first time in the present study. The length of mtDNA sequences of C. ashworthi is 13,876 bp in size, containing 12 protein-coding genes (cox1-3, nad1-6, nad4L, atp6, and cytb), two ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, and two non-coding regions. All genes are encoded by the same strand. The phylogenetic analysis by Bayesian inference method revealed that Cylicocylus genus was present in the same clade with high statistical support, and C. ashworthi was closely related with Cylicocylus nassatus than to other Strongylidae nematodes. The complete mt genome provide useful genetic marker for further studies of the taxonomy and systematics of Cyathostominae nematodes.
Cyathostominae nematodes are the most prevalent nematode pathogens with over 50 species described in equine (Lichtenfels et al. Citation2008), which inhabit in the large intestine with a high prevalence in all ages horses (van Doorn et al. 2014). Cylicocylus ashworthi is a significant Cyathostominae nematode, which can cause mild clinical symptoms such as bellyache, weight loss, and even death, leading to considerable economic losses to the livestock industry. In the present study, we assembled and characterized the complete mitochondrial genome of C. ashworthi from horse and to re-construct the phylogenetic relationships of Strongylidae nematodes.
Adult nematodes of C. ashworthi were collected from the large intestine of naturally infected horses in Daqing, Heilongjiang Province, China, which are located between east longitude of 125°10’ and north latitude of 46°53’. Individual worms were identified according to its morphological characteristics (Lichtenfels et al. Citation2008) and stored in the Department of Parasitology, Heilongjiang Bayi Agricultural University (specimen no. BYNKPL-180921). The genomic DNA was extracted from a single specimen, and the mt genome was PCR amplified utilizing primers designed according to C. nassatus previously published (Gao, Qiu, et al. Citation2017). The phylogenetic tree was constructed to study the relationships of C. ashworthi with other Strongylidae nematodes, using Bunostomum phlebotomum (accession no. FJ483517) as an outgroup.
The entire mt genome of C. ashworthi was 13,876 bp (accession no. MK422514), containing 12 protein-coding genes (PCGs, cox1-3, nad1-6, nad4L, atp6, and cytb), two ribosomal RNA (rRNA, rrnL, and rrnS) genes, 22 transfer RNA (tRNA) genes, and two non-coding regions (NCR), all genes are encoded by the same strand. Gene arrangement was identical to that of Cyathostominae nematodes mtDNA sequence belong to GA3 type (Liu et al. Citation2013). The nucleotide content of the mt genome sequence was biased toward A + T (75.4%). PCGs of C. ashworthi mt genome encoded 3418 amino acids in total. The A + T content of PCGs ranged from 69.0% (cox1) to 81.2% (nad4L). Codon usage analysis of C. ashworthi showed that two kinds of initiation codons (ATT and TTG) and three kinds of termination codons (TAA, TAG, and T) were used. The size of the 22 tRNAs ranging from 53 to 62 bp. The rrnL (978 bp) is located between trnH and nad3, whereas the rrnS (699 bp) is located between trnE and trnS(UCN). The Long NCR (270 bp) is located between trnA and trnP, which A + T content is 81.5%, and the Short NCR (82 bp) is located between nad4 and cox1, which A + T content is 79.3%.
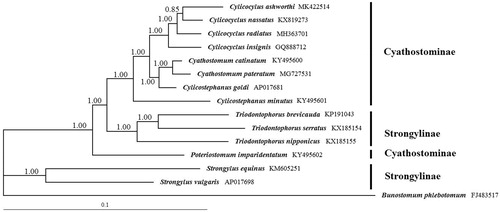
Phylogenetic analysis showed that Cylicocylus genus was present in the same clade with high statistical support, and C. ashworthi was closely related with C. nassatus than to other Strongylidae nematodes (). Notably, Triodontophorus species (T. brevicauda, T. serratus, and T. nipponicus) belong to subfamily strongylinae had relatively high identity with Cyathostominae nematodes, rather than Strongylus species (S. equinus and S. vulgaris) in the same subfamily, the results are consistent with that previously reported (Gao, Zhang, et al. Citation2017). The complete mt genome provides useful genetic marker for further studies of the taxonomy and systematics of Cyathostominae nematodes.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Lichtenfels JR, Kharchenko VA, Dvojnos GM. 2008. Illustrated identification keys to strongylid parasites (Strongylidae: Nematoda) of horses, zebras and asses (Equidae). Vet Parasitol. 156:4–161.
- Liu GH, Shao R, Li JY, Zhou DH, Li H, Zhu XQ. 2013. The complete mitochondrial genomes of three parasitic nematodes of birds: a unique gene order and insights into nematode phylogeny. BMC Genom. 14:414
- Gao Y, Qiu JH, Zhang BB, Su X, Fu X, Yue DM, Wang CR. 2017. Complete mitochondrial genome of parasitic nematode Cylicocyclus nassatus and comparative analyses with Cylicocyclus insigne. Exp Parasitol. 172:18–22.
- Gao Y, Zhang Y, Yang X, Qiu JH, Duan H, Xu WW, Chang QC, Wang CR. 2017. Mitochondrial DNA Evidence Supports the Hypothesis that Triodontophorus Species Belong to Cyathostominae. Front Microbiol. 8:1444.
- van Doorn DC, Ploeger HW, Eysker M, Geurden T, Wagenaar JA, Kooyman FN. 2014. Cylicocyclus species predominate during shortened egg reappearance period in horses after treatment with ivermectin and moxidectin. Vet Parasitol. 206:246–252.