Abstract
The Toxicodendron vernicifluum cv. Dahongpao is an elite natural triploid lacquer tree with high economic and medicinal value. Therefore, in this study, we first characterize T. vernicifluum cv. Dahongpao complete chloroplast (cp) genome sequence. The complete cp genome sequence of T. vernicifluum cv. Dahongpao, with 38.0% GC content, is 159,571 bp in length. The genome presents a typical quadripartite structure with two inverted repeat regions (each 26,511 bp in length), separated by one large single-copy region and one small single-copy region (87,475 and 19,074 bp in length, respectively). The overall GC content is 38%, while in the LSC, SSC, and IR regions the GC contents are 36.1%, 32.6%, and 43.0%, respectively. The cp genome encodes 131 functional genes, of which 114 genes are single-copy, including 80 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. Phylogenetic analysis using maximum likelihood (ML) strongly support that T. vernicifluum cv. Dahongpao has a close phylogenetic relationship with Rhus chinensis and Pistacia weinmaniifolia.
Toxicodendron vernicifluum cv. Dahongpao is a natural triploid variety of lacquer tree, which was discovered at Bashan Mountain in Shaanxi Province, China (Zhao and Wei Citation2007), and now is widely introduced into the planting area of lacquer tree. Triploid lacquer tree has higher yield of raw lacquer and stronger resistance to stress than that of diploid lacquer tree (Zhao et al. Citation2013). Chloroplast (cp) genomes are highly conserved in sequence and structure due to their non-recombinant, haploid, and uniparentally inherited nature (Birky Citation2001; Wicke et al. Citation2011). Hence, a good knowledge of its genomic information would make it easy to investigate genomic variations and contribute to further physiological molecular and phylogenetical studies of this tree species. Here, we first report the complete cp genome sequence of T. vernicifluum cv. Dahongpao using high-throughput sequencing method, and the annotated genomic sequence has been submitted to GenBank under the accession number MK550621.
The total genomic DNA was extracted with the TGuide plant genomic DNA prep kit (Tiangen Biotech, Beijing, China) from the fresh and healthy leaves of a single individual of T. vernicifluum cv. Dahongpao, which was collected from Shaanxi province, China. All the voucher specimens of T. vernicifluum cv. Dahongpao were preserved in the Herbarium of Southwest Forestry University, and DNA samples were stored at −80 °C at the Key Laboratory of State Forestry Administration on Biodiversity Conservation in Southwest China, Southwest Forestry University, Kunming, China. Total DNA was used to generate libraries with an average insert size of 350 bp with the Illumina Novaseq 6000 platform. Approximately 8 Gb raw data were produced with 150 bp pair-end read lengths. The complete cp genome assembly using GetOrganelle software (Jin et al. Citation2018) was used to assemble the complete cp genome of T. vernicifluum cv. Dahongpao, with Pistacia weinmaniifolia as the reference. Geneious R8 (Biomatters Ltd, Auckland, New Zealand) software was used for initial cp genome annotation. Start and stop codons were checked and adjusted manually when necessary by comparing to the reference genome P. weinmaniifolia.
The T. vernicifluum cv. Dahongpao cp genome is a typical circular double-stranded DNA molecule with a length of 159,571 bp. The cp genome has the usual quadripartite structure, featuring a LSC region (large single copy region, 87,475 bp), a SSC region (small single copy region, 19,074 bp), and a pair of IRs (inverted repeats, 26,511 bp). The GC contents of the LSC, SSC, IR regions, and of the complete cp genome are 36.1%, 32.6%, 43%, and 38.0%, respectively. The T. vernicifluum cv. Dahongpao cp genome encodes 131 functional genes, including 86 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Of these genes, six protein-coding genes, seven tRNA genes, and four rRNA genes within the IR regions were duplicated. We found that 15 genes (rps16, rpoC1, rpl16, rpl2, petD, petB, ndhB, ndhA, atpF, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, trnV-UAC) have one intron and three genes (ycf3, rps12, clpP) have two introns.
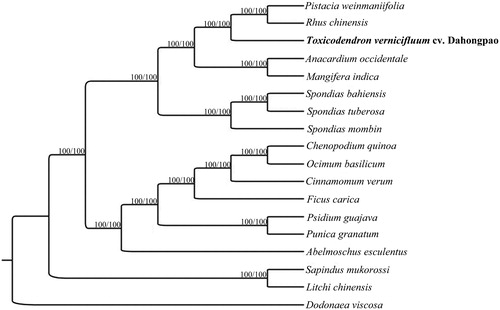
In order to perform the phylogenetic relationship of T. vernicifluum cv. Dahongpao within the family of Anacardiaceae, 14 complete cp genomes of Anacardiaceae were achieved from GenBank, and three species of Litchi chinensis, Sapindus mukorossi, and Dodonaea viscosa in the family of Sapindaceae were used as out-groups. All of these 18 complete cp sequences were aligned by the MAFFT version 7 software (Katoh and Standley Citation2013). A maximum-likelihood (ML) tree was inferred by bootstrapping with 1000 replicates through IQ-TREE 1.5.5 (Nguyen et al. Citation2015) based on the GTR + F+R4 nucleotide substitution model, which was selected by ModelFinder (Kalyaanamoorthy et al. Citation2017). The produced phylogenetic tree reveals that T. vernicifluum cv. Dahongpao is closely related to Rhus chinensis and Pistacia weinmaniifolia. ().
Figure 1. Phylogenetic relationships among 15 complete chloroplast genomes of Anacardiaceae. Bootstrap support values are given at the nodes. Chloroplast genome accession number used in this phylogeny analysis: Pistacia weinmaniifolia: MF630953; Rhus chinensis: KX447140; Toxicodendron vernicifluum cv. Dahongpao: MK550621; Anacardium occidentale: KY635877; Mangifera indica: KY635882; Spondias bahiensis: KU756561; Spondias tuberosa: KU756562; Spondias mombin: KY828469; Chenopodium quinoa: KY635884; Ocimum basilicum: KY623639; Cinnamomum verum: KY635878; Ficus carica: KY635880; Psidium guajava: KY635879; Punica granatum: KY635883; Abelmoschus esculentus: KY635876.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability
The complete chloroplast genome sequence of Toxicodendron vernicifluum cv. Dahongpao in this study were submitted to the NCBI database under the accession number MK550621.
Additional information
Funding
References
- Birky CW. 2001. The inheritance of genes in mitochondria and chloroplasts: laws, mechanisms, and models. Annu Rev Genet. 35:125–148.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assemble of a complete circular chloroplast genome using genome skimming data. bioRxiv. doi.org/10.1101/256479.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14:587–589.
- Katoh K, Standley DM. 2013. MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Wicke S, Schneeweiss GM, dePamphilis CW, Muller KF, Quandt D. 2011. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 76:273–297.
- Zhao XP, Wei SN. 2007. Genetic evaluation of Toxicodendron vernicifluum cultivars using amplified fragment length polymorphism markers. J Mol Cell Biol. 40:262–266.
- Zhao M, Liu C, Zheng G, Wei S, Hu Z. 2013. Comparative studies of bark structure, lacquer yield and urushiol content of cultivated Toxicodendron vernicifluum varieties. New Zeal J Bot. 51:13–21.
