Abstract
Yunnanilus pulcherrimus belongs to the genus Yunnanilus within the subfamily Nemacheilidae, endemic to Guangxi province, Southwestern China. In this study, we sequenced the complete mitochondrial genome of Y. pulcherrimus and submitted the genome into GenBank with accession number MK387705. The complete mitochondrial genome is 16,573 bp in length, consisting of 37 genes coding for 13 proteins, two rRNAs, 22 tRNAs, and one control region. Phylogenetic analysis using mitochondrial genomes of 16 species showed that Y. pulcherrimus was first grouped with micronemacheilus cruciatus as a monophyletic clade, but not including Y. jinxiensis.
Yunnanilus pulcherrimus, which belongs to the genus Yunnanilus within the subfamily Nemacheilidae (Teleostei, Cypriniformes), was first found in the underground river (belong to Hongshui River) of Du’an county, Southwestern China (Yang et al. Citation2004). Yunnanilus was endemic to the Yunnan Plateau, most of the species located in east-central Yunnan province, but Y. pulcherrimus only distributed in Guangxi province (Chen Citation2013; Jiang et al. Citation2016). Previous morphological studies showed that this species belonged to Y. pleurotaenia group (Yang et al. Citation2004); however, there is a lack of molecular proof. Here, we first determined the complete mitochondrial genome of Y. pulcherrimus and analyzed the phylogenetic relationship of the loach in Nemacheilidae fishes, contributing to aid further genetic and evolutionary researches of this fish species.
The sample of Y. pulcherrimus was collected from the Dule river (belong to Liujiang River) located in Liuzhou City of Guangxi, China (24°13′44.26′N, 109°25′16.24′E), and its muscle samples were fixed in 95% ethanol and preserved in Liuzhou Aquaculture Technology Extending Station, Liuzhou, China. Total genomic DNA was extracted from the fins through using traditional phenol-chloroform extraction method (Taggart et al. Citation1992). Then, we have sequenced the complete mitochondrial genome of Y. pulcherrimus using the Illumina Hiseq4000 platform with de novo strategy (Tang et al. Citation2015) and submitted the genome into GenBank with accession number MK387705.
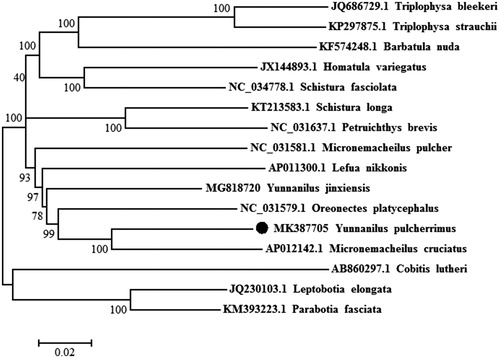
The circular mitogenome of Y. pulcherrimus was 16,573 bp in length, including 13 protein-coding (PCGs), two ribosomal RNA (rRNA), 22 transfer RNA (tRNA) genes, and one non-coding control region (D-loop). According to BLAST comparison results, Y. pulcherrimus was the most similar to Micronemacheilus cruciatus (synonymy: Yunnanilus cruciatus), showing 89–95% identities to Micronemacheilus cruciatus by compared with the whole mitogenome and 16S rRNA sequences, respectively. The gene arrangement and structure were identical to the most vertebrates as previously reported, and the nucleotide base composition of Y. pulcherrimus was 29.88% A, 16.53% G, 26.10% T, and 27.50% C, and the overall GC and AT content were 44.02% and 55.98%, respectively. To validate the phylogenetic position of Y. pulcherrimus, a neighbour-joining (NJ) tree inferred from the whole mitogenome of 15 Nemacheilidae species and 1 Cobitidae species (outgroup) was constructed with 1000 bootstrap replicates using MEGA 6.06. The phylogenetic results revealed that Y. pulcherrimus was first grouped with micronemacheilus cruciatus as a monophyletic clade (), but not including Y. jinxiensis which also belonged to the same genus Yunnanilus (Huang et al. Citation2019). It suggested that morphological classification methods in the genus Yunnanilus were not necessarily scientific, and more molecular data is needed to prove it.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Chen XY. 2013. Checklist of fishes of Yunnan. Zool Res. 34:281–343.
- Huang W, Shi GF, He AY, Wen YH, Huang J, Luo FG. 2019. Characterization of complete mitochondrial genome of Yunnanilus jinxiensis (Cypriniformes, Balitoridae, Nemacheilidae). Mitochondrial DNA B. 4:185–186.
- Jiang ZG, Jiang JP, Wang YZ, Zhang E, Zhang YY, Li LL, Xie F, Cai B, Cao L, Zheng GM, et al. 2016. Red list of China’s vertebrates. Biodivers Sci. 24:500–551.
- Taggart J, Hynes R, Prod Uhl P, Ferguson A. 1992. A simplified protocol for routine total DNA isolation from salmonid fishes. J Fish Biol. 40:963–965.
- Tang M, Hardman CJ, Ji Y, Meng G, Liu S, Tan M, Yang S, Moss ED, Wang J, Yang C, et al. 2015. High-throughput monitoring of wild bee diversity and abundance via mitogenomics. Methods Ecol Evol. 6:1034–1043.
- Yang JX, Chen XY, Lan JH. 2004. Occurrence of two new Plateau-indicator loaches of Nemacheilinae (Balitoridae) in Guangxi with reference to zoogeographical significance. Zool Res. 25:111–116.