Abstract
Cophecheilus bamen is a freshwater cyprinid fish that endemic to Jingxi and Tiandong county, Southwestern China. In this study, we first determined the complete mitochondrial genome sequence of C. bamen. The circular mitogenome was 16,588 bp in length and consisted of 13 protein-coding genes, two ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, and one non-coding control region. Phylogenetic analysis using mitochondrial genomes of 16 species showed that C. bamen is supported as a single monophyletic clade and sister to the Parasinilabeo, Pseudogyrinocheilus, Semilabeo, Discogobio, Pseudocrossocheilus (synonymy: Sincrossocheilus), and Ptychidio genus.
Cophecheilus bamen, a freshwater cyprinid fish, was first found in a tributary of the Zuo-Jiang of the Pearl River drainage in Jingxi County, Guangxi Province, South China (Zhu et al. Citation2011). As a new genus, Cophecheilus had the unusual oromandibular structures by compared with all other Asian garrains; and, so far, this genus only had two effective species (C. brevibarbatus and C. bamen), both of them were endemic to Guangxi province of China (Jiang et al. Citation2016). Here, we determined the complete mitochondrial genome sequence of C. bamen and analyzed the phylogenetic relationship of the Labeonini fishes, contributing to aid further genetic and evolutionary researches of this fish species.
The sample of C. bamen was collected from the underground river located in Tiandong County of Guangxi, China (23°32′28.33′N, 107°11′11.62′E), and its muscle samples were fixed in 95% ethanol and preserved in Liuzhou Aquaculture Technology Extending Station, Liuzhou, China. Total genomic DNA was extracted from the fins through using traditional phenol-chloroform extraction method (Taggart et al. Citation1992). Then, we have sequenced the complete mitochondrial genome of Y. jinxiensis using the Illumina Hiseq4000 platform with de novo strategy (Tang et al. Citation2015), and submitted the genome into GenBank with accession number MK387703.
The circular mitogenome of C. bamen was 16,588 bp in length, including 13 protein-coding (PCGs), two ribosomal RNA (rRNA), 22 transfer RNA (tRNA) genes, and one non-coding control region (D-loop). The overall base composition was 32.26% A, 25.87% T, 26.51% C, and 15.36% G with 41.87% GC content. All genes were encoded on the heavy strand but ND6 gene and eight tRNA genes. The regular initiation codon ATG, which appeared in 13 PCGs except in COI gene, has GTG as the start codon. Furthermore, two complete types (TAA, TAG) of the stop codon appeared in six genes of 13 PCGs (ND1, ND5, ND6, ND4L, COII, and ATP8), whereas the two incomplete types (TA-, T–) appeared in the other seven genes. The length of 22 tRNAs ranged from 67 bp to 76 bp in size, and the 12S and 16S rRNA genes were 952 bp and 1,687 bp in length, respectively. The D-loop was located between tRNA-Phe and tRNA-Pro with an AT bias of 68.53%.
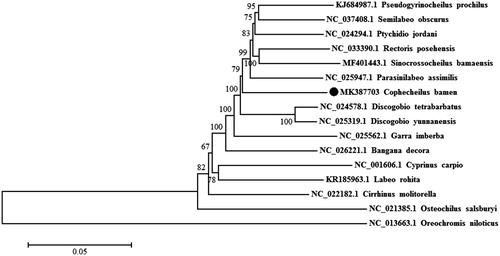
To validate the phylogenetic position of C. bamen, a neighbour-joining (NJ) tree inferred from the whole mitogenome of 15 Labeonini fishes and one Cichlidae species (outgroup) was constructed with 1000 bootstrap replicates using MEGA 6.06. The phylogenetic results revealed that C. bamen is supported as a single monophyletic clade and sister to the Parasinilabeo, Pseudogyrinocheilus, Semilabeo, Discogobio, Pseudocrossocheilus (synonymy: Sincrossocheilus), and Ptychidio genus (); interestingly, C. bamen and its close relationships species all have the similar oromandibular structures.
References
- Jiang ZG, Jiang JP, Wang YZ, Zhang E, Zhang YY, Li LL, Xie F, Cai B, Cao L, Zheng GM, et al. 2016. Red list of China’s vertebrates. Biodivers Sci. 24:500–551.
- Taggart J, Hynes R, Prod Uhl P, Ferguson A. 1992. A simplified protocol for routine total DNA isolation from salmonid fishes. J Fish Biol. 40:963–965.
- Tang M, Hardman CJ, Ji Y, Meng G, Liu S, Tan M, Yang S, Moss ED, Wang J, Yang C, et al. 2015. High-throughput monitoring of wild bee diversity and abundance via mitogenomics. Methods Ecol Evol. 6:1034–1043.
- Zhu Y, Zhang E, Zhang M, Han YQ. 2011. Cophecheilus bamen, a new genus and species of labeonine fishes (Teleostei: Cyprinidae) from South China. Zootaxa. 2881:39–50.