Abstract
Nitraria tangutorum and N. roborowskii are the representative species of Nitraria and mainly distributed in the salt deserts regions of northwest China. In the current study, we report the complete sequences of the chloroplast genomes of N. tangutorum and N. roborowskii, which were 159,383 and 159,397 bp in length, respectively, with 112 predicted genes consisting of 79 protein-coding genes, 39 tRNA genes, and eight rRNA genes. The GC content of the two chloroplast genomes were 37.15% and 37.32% in N. tangutorum and N. roborowskii, respectively. ML phylogenomic analysis indicated that N. tangutorum and N. roborowskii formed one monophyletic group as the basal position of Sapindales clade and supporting separate this genus form Zygophyllaceae.
Nitraria L. is a small genus including about 11 species and mainly distributed in salt deserts, salt marshes, and coastal sand dunes regions of Africa, Europe, Asia, and Australia (Kubitzki Citation2011). There are about six species distributed in China, whereas Nitraria tangutorum and N. roborowskii is widely distributed in the northwest region of China (Wu et al. Citation2008). As an advantageous shrub for windbreak and sand fixation, N. tangutorum and N. roborowskii have an important role in ecological environment protection (Luo et al. Citation2008). Meanwhile, they are also used as a traditional Tibetan medicine and important nutritional and health food resources (Yang Citation1991). Nitraria was first placed in Zygophyllaceae (Pan et al. Citation1999). However, it differs from the Zygophyllaceae in many morphological characters (Nurbay and Pan Citation2003) and molecular evidence also supports its recognition as a separate family named Nitrariaceae (APG 2009). To date, there is still a controversy about the phylogenetic status of Nitraria with the relative genus and need to be further elucidated.
In the current study, we report the completed chloroplast genomes of N. tangutorum and N. roborowskii collected in Qinghai Province, China, and the specimens were deposited in the Qinghai-Tibetan Plateau Museum of Biology (HNWP). The DNA was isolated from fresh leaves via the modified CTAB method (Doyle Citation1987). The complete chloroplast genome was sequenced using Illumina HiSeq2500 platform, assembled with SPAdes (Bankevich et al. Citation2012), and annotated with CpGAVAS (Liu et al. Citation2012). The two completed chloroplast genome sequences together with 15 species from Sapindales, two species from Zygophyllaceae and Nicotiana tabacum as outgroup were aligned with MAFFT (Katoh and Standley 2013). Gblocks (Castresana Citation2000) was employed to remove ambiguously aligned sites. A maximum likelihood (ML) analysis was conducted using RAxML-HPC2 on XSEDE based on the GTR + G + I nucleotide substitution model with 1000 replications.
The two complete chloroplast genomes of Nitraria have a total length of 159,397 bp for N. roborowskii (MK347421) and 159,383 bp for N. tangutorum (MK347423). The typical quadripartite structure consisted of a pair of IRs of 26589 bp and 26,586 bp, an LSC region of 87,907 bp, and 87,901 bp, an SSC region of 18,312 bp and 18,310 bp in N. roborowskii and N. tangutorum, respectively. The GC content of the two chloroplast genomes were 37.32% and 37.15%. There were 112 predicted genes in both of the two species consisting of 79 protein-coding genes, 39 tRNA genes, and eight rRNA genes. Among all the genes, 16 genes contained introns, and 13 of them (six protein-coding genes, six tRNA genes, and one rRNA genes) have one intron, whereas two (clpP and ycf3) have two introns.
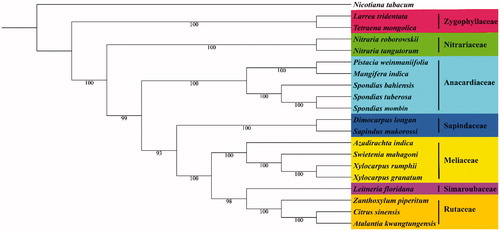
ML analysis showed that N. tangutorum and N. roborowskii constituted one monophyletic group as Nitrariaceae (). Additionally, Nitrariaceae was in the basal position of Sapindales clade and showed closed relationship with Anacardiaeceae, and Spindaceae, rather than the Zygophyllaceae. These newly reported chloroplast data could be useful in taxonomic study of this genus, and of high values for the genetic engineering or conservation genetics involving these species.
Figure 1. Maximum likelihood phylogenetic tree based on 20 complete chloroplast genome sequences. The number on each node indicates the bootstrap value. Accession numbers: Nitraria tangutorum MK347423, Nitraria roborowskii MK347421, Larrea tridentate KT272174.1, Tetraena mongolica MH325022.1, Zanthoxylum piperitum KT153018.1, Citrus sinensis DQ864733.1, Azadirachta indica KF986530.1, Sapindus mukorossi KM454982.1, Leitneria floridana KT692940.1, Pistacia weinmaniifolia MF630953.1, Dimocarpus longan MG214255.1, Spondias tuberosa KU756562.1, Spondias bahiensis KU756561.1, Spondias mombin KY828469.1, Atalantia kwangtungensis MH329190.1, Mangifera indica KY635882.1, Xylocarpus granatum MH348155.1, Xylocarpus rumphii MH330687.1, Swietenia mahagoni MH348156.1, and Nicotiana tabacum Z00044.2.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552.
- Doyle JJ. 1987. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kubitzki K, editor 2011. The families and genera of vascular plants Vol. X. Flowering plants. Eudicots: Sapindales, Cucurbitales, Myrtaceae. Berlin: Springer Nature Switzerland AG.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and genbank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715
- Luo G, Zhou C, Chen X, Li Y. 2008. A methodology of characterizing status and trend of land changes in oases: a case study of Sangong River watershed, Xinjiang, China. J Environ Manage. 88:775–783.
- Nurbay A, Pan XL. 2003. Pollen morphology and taxonomy of Nitraria and its allied genera in West China. Arid Zone Res. 20:16–21.
- Pan XL, Shen GM, Chen P. 1999. A preliminary research on taxonomy and systematics of genus Nitraria. Acta Botanica Yunnanica. 21: 287–295.
- The Angiosperm Phylogeny Group (APG). 2009. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc. 161:105–121.
- Wu ZY, Raven PH, Hong DY. 2008. Flora of China: Vol. 11. St. Louis (MO): Missouri Botanical Garden.
- Yang YC. 1991. Flora Tebitan Medicine. Xininng (China): Qinghai People's Publishing House.
