Abstract
Trichiurus is one of the most important economic fishes around the world. In the present study, we determined the complete mitochondrial genome of Trichiurus sp. from Jiangsu is a circular-molecule of 16,662 bp in length, which encodes 35 genes in all. These genes comprise 13 protein-coding genes, 20 transfer RNA genes, and two ribosomal RNA genes, with gene arrangement and content basically identical to those of other species of Trichiuridae. The result of phylogenetic analysis strongly supported that Trichiurus sp. should be Trichiurus japonicus.
Trichiurus, one of the Trichiuridae, exists many geographical populations, is a species complex that includes several groups being in the Atlantic Ocean, East Pacific Ocean, Northwest Pacific Ocean and Indo-Pacific Ocean (Chakraborty et al. Citation2006; Tzeng et al. Citation2007; Burhanuddin and Parin Citation2008; Shih et al. Citation2011). In China, it is distributed over the Yellow Sea, East China Sea, White Sea, and South China Sea. It is one of the most important economic fishes, and known as China's four largest seafood with large yellow croaker (Pseudosciaena crocea), small yellow croaker (Larimichthys polyactis), and cuttlefish (Sepiella maindroni). Until now, we cannot identify the Trichiurus species by using morphology. Thus, we determined the complete mitochondrial DNA sequence and organization of Trichiurus sp. from Jiangsu here.
The complete mitogenome sequence of Trichiurus sp. was determined using 14 pairs of primers which were designed based on the mitogenome sequence of Trichiurus japonicus (GenBank number NC 011719.1). The specimen of Trichiurus sp. was obtained from Yangkou Port, Qidong, Jiangsu, China (124°48′17″E, 30°58′39″N), and the whole muscle was immediately preserved in 95% ethanol. Total genomic DNA was extracted using a standard phenol-chloroform method (Sambrook et al. Citation1989) and stored at −20 °C in Laboratory of Aquatic Health and Public Health, College of Animal Science and Technology, Anhui Agricultural University, Anhui, China.
The complete mitogenome of Trichiurus sp. was determined to be 16,662 bp (GenBank accession number MH037011), which comprises 35 coding and two non-coding regions. The 35 coding regions include 13 protein-coding genes (ATP 6 and 8, COX I-III, Cytb, ND 1-6, and 4L), 20 transfer RNA genes (tRNAs), and two ribosomal RNA genes (12S and 16S rRNAs), and two non-coding regions consist of light-strand replication origin (OL) and control region (CR). Except for one protein-coding gene ND 6 and seven tRNAs (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, and tRNAGlu), all other genes are encoded on the heavy strand, which are in accordance with the other teleost mitogenomes (Oh et al., Citation2007; Zhu et al. Citation2013).
All protein-coding genes in Trichiurus sp. have a methionine (ATG) start codon except for ND 1, COX I and ND 6, which starts with ATA, ATA and AAC. The 13 protein-coding genes are ended with AAC, ATT, ATG, ACT, AGT, TTG, TCT, AAG, TGT, CTA, CTA, TAC, and CAA.
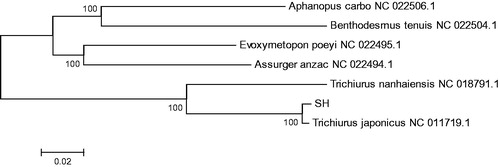
The phylogenetic analysis was performed by MEGA 6.06 program (Tamura et al., Citation2013) based on the complete mitogenome sequence of Trichiurus sp. and those of 6 closely related species belonging to five genus Aphanopus, Assurger, Benthodesmus, Evoxymetopon, and Trichiurus. The neighbor-joining tree () showed that Trichiurus sp. first clustered together with T. japonicus and formed a monophyly in the genus Trichiurus, and then they constituted a sister-group relationship with other four genus. The present results on the molecular phylogenetic analysis strongly supported that the Trichiurus sp. should be T. japonicus. This study also revealed the phylogenetic relationship of the Trichiuridae at molecular levels.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Burhanuddin AI, Parin NV. 2008. Redescription of the Trichiurid fish, Trichiurus nitens, Garman, 1899, being a valid of species distinct from T. lepturus, Linnaeus, 1758 (Perciformes: Trichiuridae). J Ichthyol. 48:825–830.
- Chakraborty A, Aranishi F, Iwatsuki Y. 2006. Genetic differentiation of Trichiurus japonicus and T. lepturus (Perciformes: Trichiuridae) based on mitochondrial DNA analysis. Zool Stud. 45:419–427.
- Oh DJ, Kim JY, Lee JA, Yoon WJ, Park SY, Jung YH. 2007. Complete mitochondrial genome of the rock bream Oplegnathus fasciatus (Perciformes, Oplegnathidae) with phylogenetic considerations. Gene. 392:174–180.
- Sambrook J, Maniatis TE, Fritsch EF. 1989. Molecular cloning: a laboratory manual. Vol 1. New York: Cold Spring Harbor Laboratory Press.
- Shih NT, Hsu KC, Ni IH. 2011. Age, growth and reproduction of cutlassfishes Trichiurus spp. in the Southern East China Sea. J Appl Ichthyol. 27:1307–1315.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. Mega6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Tzeng CH, Chen CS, Chiu TS. 2007. Analysis of morphometry and mitochondrial DNA sequences from two Trichiurus species in waters of the Western North Pacific: taxonomic assessment and population structure. J Fish Biol. 70:165–176.
- Zhu YX, Chen Y, Cheng QQ, Qiao HY, Chen WM. 2013. The complete mitochondrial genome sequence of Schizothorax macropogon (Cypriniformes: Cyprinidae). Mitochondrial DNA. 24:237–239.