Abstract
Cassia tora (Senna tora), known as an economically important plant, is medicinal in nature and belongs to the Fabaceae family. The complete mitochondrial genome sequences of S. tora were 566,589 bp in length with a 45.23% GC content. A total of 63 genes were annotated including 36 protein-coding genes, 22 tRNA genes, and 5 rRNA genes. Phylogenetic tree based on the mitochondrial genome demonstrated that S. tora was most closely related to the Senna occidentalis and Caesalpinioideae subfamily and is definitely separated from the Faboideae subfamily.
Cassia tora (Senna tora), a medicinal plant belonging to the Fabaceae family, grows all over the tropical countries. The plant was used traditionally to cure diabetes, dermatitis, constipation, cough, cold, and fever (Kumar and Roy Citation2018). A sample was obtained from the National Agrobiodiversity Center (voucher number: IT89788, geographic coordinate: 35° 49′ 19′ N, 127° 8′ 56′ E) in Jeonju, Korea. The whole genome sequencing was performed using a hybrid data of Illumina (Hiseq4000, Illumina, CA, USA) and PacBio RSII platform (Pacific Biosciences, Menlo Park, CA). First, initial contigs were generated using de novo assembly of Illumina Paired-End data and were compared with mitochondrial genome references. Second, selected contigs were performed to gap-filling procedures and error-corrected using PacBio data. Finally, genes of the mitochondrial genome were annotated using Mitofy program and BLAST searches (Alverson et al. Citation2010).
The complete mitochondrial genome of S. tora was a circular form of 566,589 bp in length with a 45.23% GC content. The genome harbored 63 annotated genes, including 36 protein-coding genes, 22 tRNA genes, and 5 rRNA genes. The annotated protein-coding sequence (exon only) accounts for 6.0% of the genome and an average gene length was 35,750 bp (including introns). The complete mitochondrion of S. tora (Accession no. MF358693) was registered to NCBI GenBank.
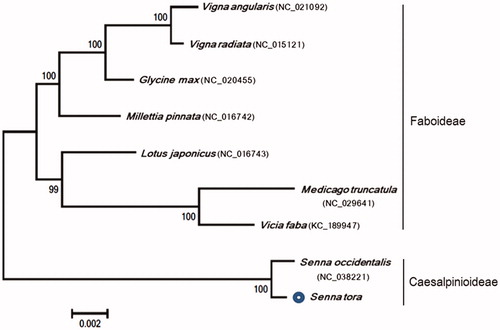
Phylogeny of S. tora to other species in the Fabaceae family was generated. Phylogenetic relationships of 21 common protein-coding genes (i.e. atp4, atp6, atp8, atp9, ccmB, ccmC, ccmFn, cob, cox1, cox3, matR, nad2-4, nad6-7, nad9, rpl16, rps3, rps12, and rps14) in S. tora were revealed by comparing the data with the eight reported species in the Fabaceae family. Genome alignments were performed using CLC Genomics Workbench 6.5 (CLC Inc., Aarhus, Denmark), and a phylogenetic tree was constructed using MEGA 7.0 (Kumar et al. Citation2016). The phylogeny indicated that S. tora species is most closely related to the Senna occidentalis of Fabaceae family, and S. tora (subfamily: Caesalpinioideae) is definitely separated from the subfamily Faboideae (). This study will contribute to developing molecular markers for germplasm classification and S. tora breeding.
Disclosure statement
The authors report no conflicts of interest.
Additional information
Funding
References
- Alverson AJ, Wei X, Rice DW, Stern DB, Barry K, Palmer JD. 2010. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol Biol Evol. 27:1436–1448.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Kumar V, Roy BK. 2018. Population authentication of the traditional medicinal plant Cassia tora L. based on ISSR markers and FTIR analysis. Sci Rep. 8:10714.